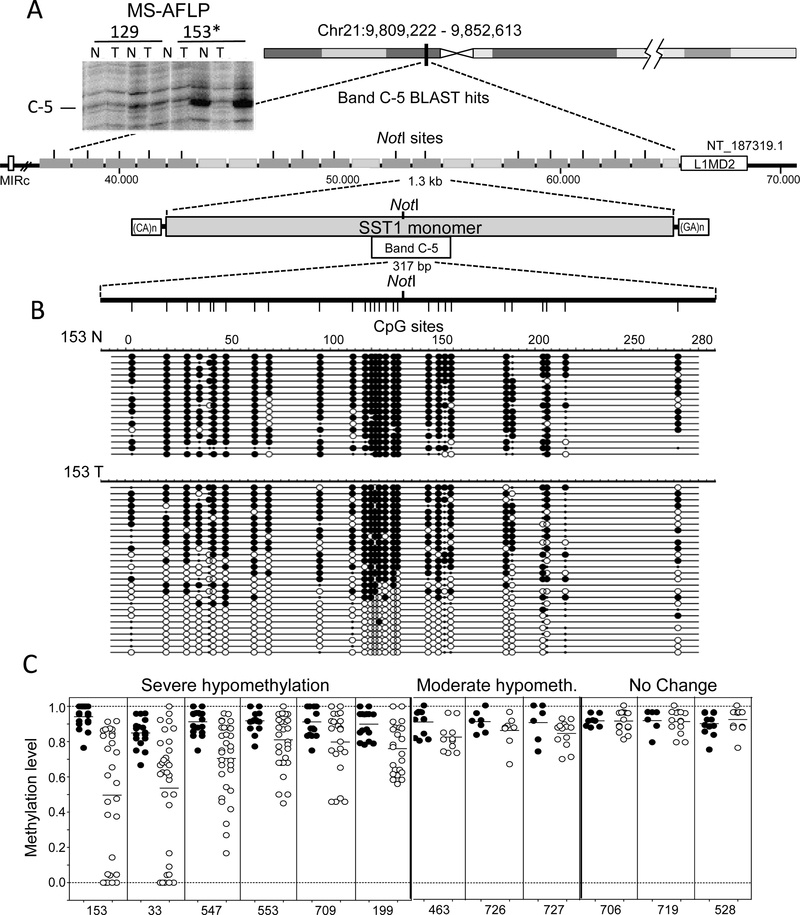
Figure 1.
SST1 elements are demethylated in colorectal cancer. (A) DNA fingerprinting by methylation sensitive-amplified fragment length polymorphism (MS-AFLP) identified a hypomethylated region in 22% of colorectal cancer (CRC) tumors (band C-5). The insert depicts the region of the fingerprint of two CRC cases, showing the intense bandC-5 in tumor 153, indicating demethylation of the NotI site. Band C-5 correspond (bed to a pericentromeric region on chromosome 21, which contains 21 SST1 satellite tandem repeats. Most of these SST1 elements contain a NotI site; that can be detected by MS-AFLP. Each repeat of approximately 1.3 kb (grey rectangles) is flanked by CA- and GA-rich regions. On each side of the21 repeat region, families of retrotransposons are present (MIRC and LIM2). N and T: normal and tumor tissue DNA; (B) A region of 317 bp comprising 28 CpG sites and the NotI site was analyzed by bisulfite sequencing in several CRCs. Analysis of case 153 is represented, where black and white circles represent methylated and unmethylated CpG sites, respectively. Small black dots indicate polymorphisms including TpA sequences possibly resulting from spontaneous deamination of methylated CpGs on the opposite DNA strand. Each row represents the sequence of an individual plasmid clone; and (C) methylation status of SST1 satellites from chromosome 21 in 12 CRCs cases. SST1 methylation levels indicate the methylation average of the 28 CpGs analyzed by bisulfite sequencing from individual PCR clones (rows in (B)) in colon normal samples (black) and tumor samples (white). Severe demethylated cases show a methylation average difference between normal and tumor equal or higher than 10%, whereas moderate demethylation cases show a difference between 5% and 10%.