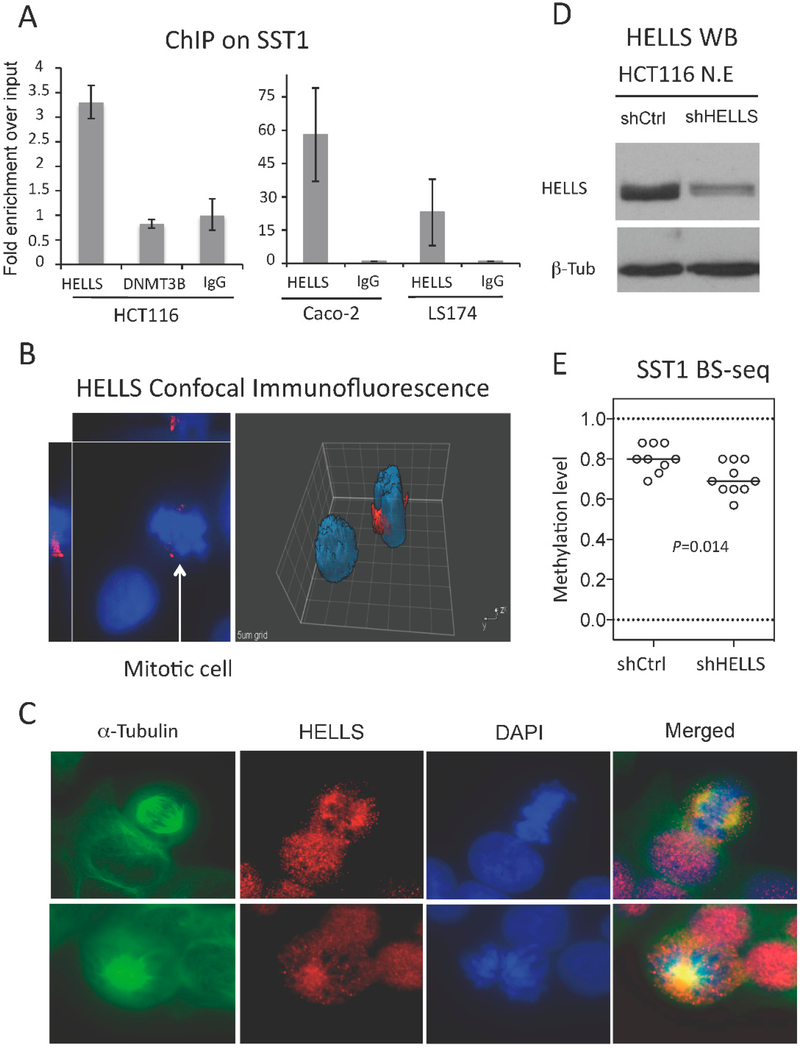
Figure 6.
HELLS is recruited to SST1 regions, localizes in the centrosome and the mitotic spindle, and HELLS knockdown results in SST1 demethylation. (A) ChIP analysis of HELLS and DNA methyltransferase 3B (DNMT3B) recruitment to SST1 repetitive elements in CRC cell line HCT116 (left graph). The right graph shows HELLS recruitment to SST1 monitored by ChIP in Caco-2 (SST1 methylated) and LS174 (SST1 demethylated). Relative recruitment was calculated by the comparative Ct method and the input values were used as normalizers. IgG shows art immunoprecipiation background; (B) HELLS expression by confocal immunofluorescence. Caco-2 cells stained with primary antibody against HELLS (red) and counterstained with 4′,6-diamidino-2-phanylindole (DAPI) (blue). Image analysis by Slidebook software v4 (Intelligent Imaging Innovations, Denver, CO, USA) shows increased protein levels of HELLS around what seems to be the centrosome regions of mitotic cells; (C) Co-immunofluorescence of HELLS with α-tubulin, a marker of the mitotic spindle, in HCT116 cells; (D) Western blot on nuclear extracts from HCT116 polyclones obtained after lentiviral infection of small hairpin RNAs (shRNA) of a scrambled control sequence (shCtrl) or targeting HELLS (shHELLS); and (E) SST1 methylation levels of individual clones analyzed by bisulfite sequencing in colon cancer cell line HCT116 with scrambled shRNA (shCtrl) or shRNAs for HELLS (shHELLS). This analysis was performed three weeks post-infection. p value was calculated by Mann Whitney test.