Figure 2.
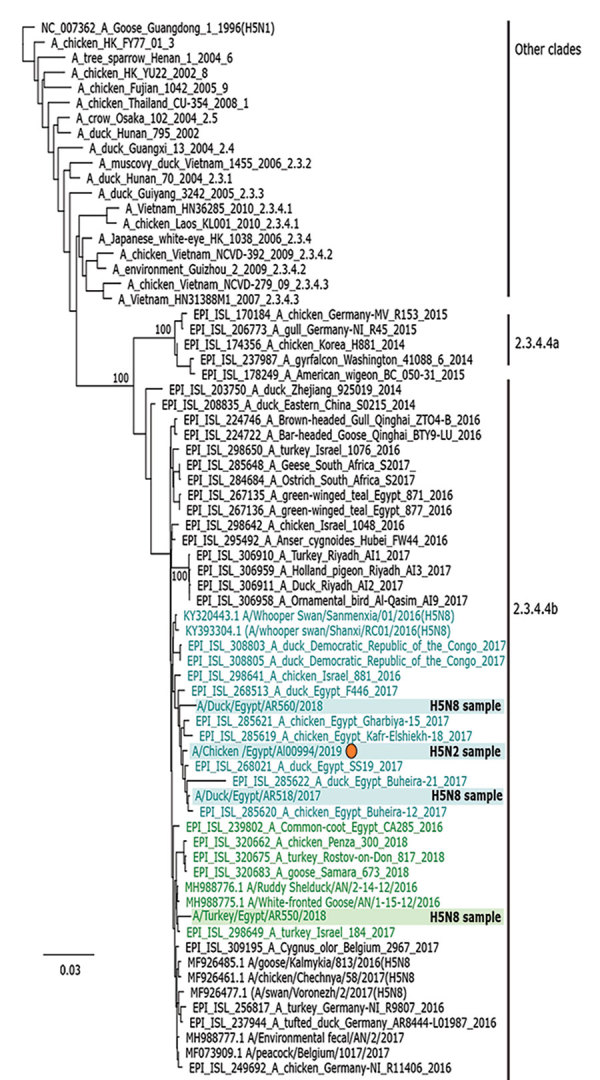
Phylogenetic analysis of the hemagglutinin segments of reassortant highly pathogenic avian influenza H5N2 and H5N8 viruses belonging to clade 2.3.3.4b from Egypt and reference viruses. Sequence analysis was based on alignment analyses by MAFFT version 7.450 embedded in the Geneious software suite, version 11.1.7 (https://www.geneious.com) with manual editing. We performed maximum-likelihood calculations using PhyML version 3.0 (http://www.atgc-montpellier.fr/phyml); we chose the best-fit model according to the Bayesian selection criterion using Model Finder embedded in Geneious. Colors indicate grouping of segment origin and match those shown in Figure 1: blue, most closely related to H5N8 viruses from China; green, most closely related to H5N8 viruses from Russia and Europe. GenBank or GISAID accession numbers (http://www.gisaid.org) are provided for reference sequences.
