Fig. 2. Frameshifting effect of mutations at the tip of domain 4 of EF-G.
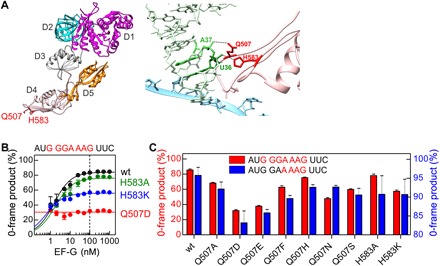
(A) Left: Crystal structure of EF-G [Protein Data Bank (PDB): 3J5X]. Domains (D) 1 to 5 and the positions of residues Q507 and H583 (red) in domain 4 are indicated. Right: Interaction of EF-G residues Q507 and H583 with the tRNA in the ap/P state of the ribosome (PDB: 4V5F). (B) Fraction of the 0-frame peptide depending on the concentration of wt EF-G and EF-G mutants. Lines are visual guides, and the dashed line indicates the concentration at which ribosomes and EF-G are equimolar. Error bars represent SD of three experiments (n = 3). (C) Effect of different substitutions of Q507 and H583 in EF-G on the fraction of 0-frame product. Error bars represent SD of three experiments (n = 3).
