Fig. 5. MTIF3 does not affect mitochondrial ribosome assembly but alters the interactions between mitoribosomes and mt-mRNAs.
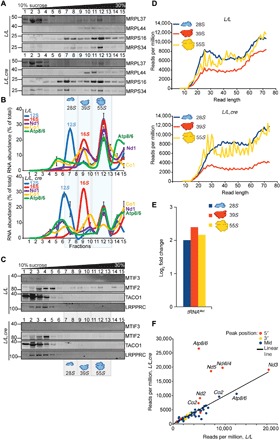
(A) Mitochondrial ribosome subunits and translating ribosomes were resolved on 10 to 30% sucrose gradients in control (L/L) and knockout (L/L, cre) 25-week-old mice. MTIF2 and the mitochondrial ribosomal protein markers of the small (MRPS34 and MRPS16) and large (MRPL44 and MRPL37) ribosomal subunits were detected by immunoblotting. The data are representative of results from four independent biological experiments. (B) The distributions of the 12S and 16S rRNAs and mRNAs in sucrose gradients were analyzed by qRT-PCR. The data are expressed as a percentage total of RNA abundance and show results from three independent biological replicates. (C) Mitochondrial ribosome subunits and translating ribosomes were resolved on 10 to 30% sucrose gradients in control (L/L) and knockout (L/L, cre) 25-week-old mice. MTIF3, MTIF2, TACO1, and LRPPRC were detected by immunoblotting. The data are representative of results from four independent biological experiments. (D) The normalized read length profile (reads per million) of the mitoribosome and its subunits on mRNAs is altered in the absence of MTIF3. (E) The association of tRNA-Met with the mitoribosome and its subunits is increased in the absence of MTIF3. (F) Enriched peaks at the 5′ end of mt-Atp8/6, mt-Nd4l/4, and mt-Nd5 mRNAs associating with the translating ribosome indicate ribosomal stalling. Peaks were considered 5′ or 3′ if they partially or fully overlapped a 100-nt window centered on the transcript 5′ or 3′ ends.
