Fig. 6. The mechanism of MTIF3 and its role in mRNA recognition and translation initiation.
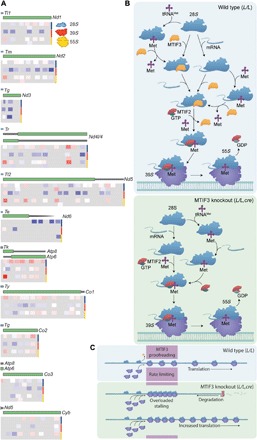
(A) Mitochondrial ribosome profiling shows the distribution of footprints on mt-mRNAs in the SSU (first two tracks), large subunit (middle two tracks), and mitoribosome (bottom two tracks) that are enriched (red) or depleted (blue) in the knockout compared to control mice. Double tracks per ribosomal fraction were used to distinguish partially overlapping footprints on the mRNAs. (B) Schematic showing the proposed roles of MTIF3 in mitochondrial translation (wild type, L/L). MTIF3 prevents the translation initiation complex formation if it is bound by a tRNA in the absence of mRNA. Only small ribosomal subunits that have bound mRNA before the recruitment of tRNA and MTIF2 are able to proceed from translation initiation to elongation. In the absence of MTIF3 (MTIF3 knockout, L/L,cre) preinitiation complexes cannot remove tRNAMet and positioning of the mRNA and its start codon cannot be monitored, so these complexes are able to participate in mature initiation complex formation. GDP, guanosine diphosphate; GTP, guanosine triphosphate. (C) Without the molecular proofreading steps performed by MTIF3, translation initiation proceeds at an accelerated rate but at the expense of fidelity. When fidelity of initiation is compromised, initiation complexes can stall at the 5′ ends of mRNAs, leaving the remainder of the mRNA prone to degradation by 3′-5′ exoribonucleases.
