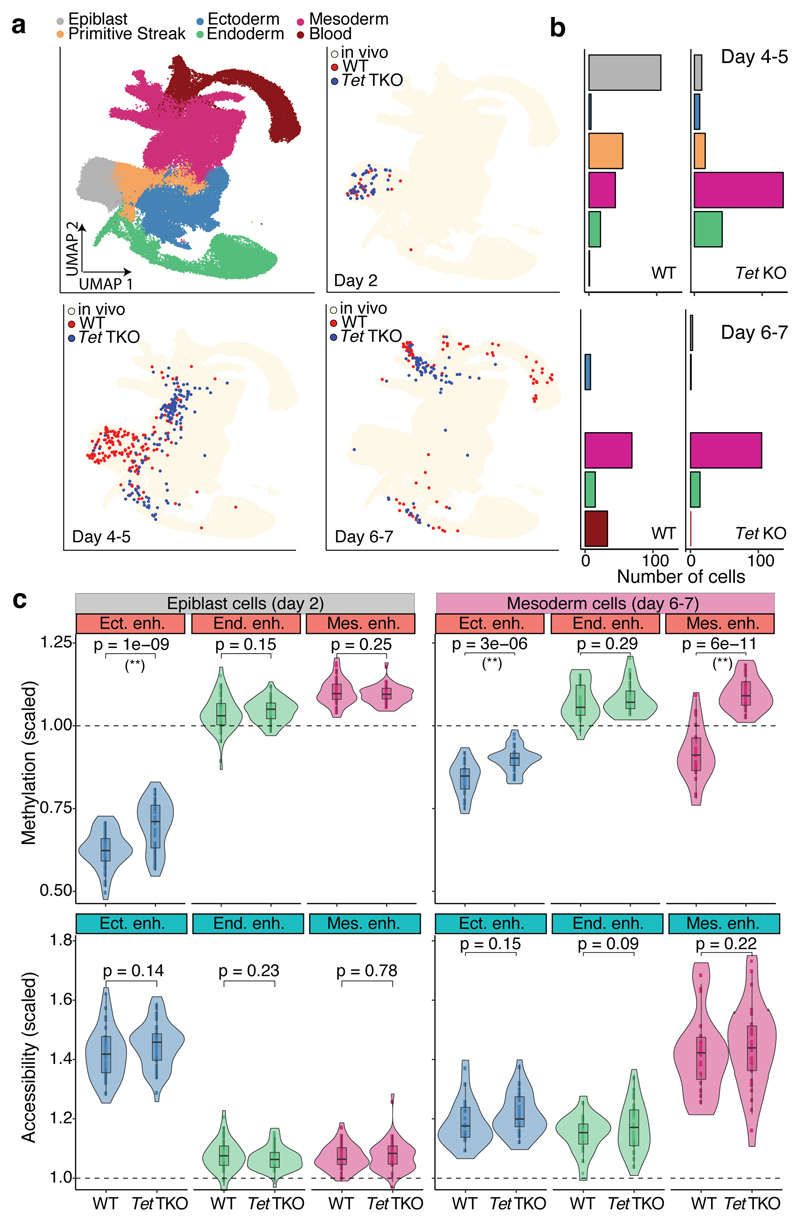
Fig. 4. TET enzymes are required for efficient demethylation of mesoderm-defining enhancers and subsequent blood differentiation in embryoid bodies.
a, UMAP projection of stages E6.5 to E8.5 of the atlas data set (no extraembryonic cells). In the top left plot cells are coloured by lineage assignment. The remaining plots show, for different days of EB differentiation, the nearest neighbours that were used to assign cell type labels to the EB data set. WT cells are red (n=438),Tet TKO cells are blue (n=436). We grouped days 4-5 and 6-7 together due to similarity in the cell types recovered. b, Bar plots showing the cell type numbers for each day of EB differentiation, grouped by genotype. c, Overlaid box and violin plots display the distribution of DNA methylation (top) or chromatin accessibility values (bottom) for lineage-defining enhancers in epiblast-like cells at day 2 (n=46 for WT and n=44 TKO) and mesoderm-like cells at days 6-7 (n=22 for WT and n=32 TKO). The y-axis shows the methylation or accessibility (%) scaled to the genome-wide levels. P-values resulting from comparisons of group means (t-test) are displayed. Asterisks denote significant differences (FDR<10%).