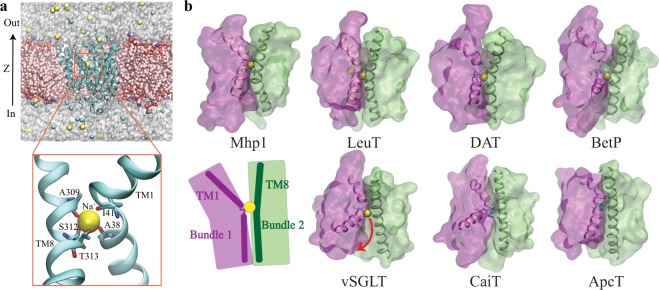
Figure 1.
A conserved ion-binding site (Na2 site) in LeuT-fold transporters. (a) Overview of the structure of Mhp1 and the simulation system. (top) The simulation system. Mhp1 is rendered in cartoon, with the bound Na+ drawn in VDW. The POPE lipids and the solute ions are also drawn in VDW, and the water molecules are represented as semi-transparent surfaces. Lipids molecules overlapping with the protein are hidden for clarity. (bottom) A close view of the highly conserved Na+-binding site (Na2). Residues surrounding the Na+-binding site along with helices TM1 and TM8 are shown and labeled. (b) The Na2 site is a conserved motif shared by Na+-dependent transporters, namely, Mhp1, LeuT, DAT, BetP and vSGLT, and is replaced by a basic residue with a similar role in two Na+-independent transporters: CaiT and ApcT. The top panel shows four transporters in their OF states, with a well-coordinated Na2 site, while the bottom panel shows three transporters in either IF or occluded state with a more open Na2 site. TM1 and TM8 are represented in cartoon form, with the bound Na+ drawn in VDW. The two helical bundles are shown in transparent surface representation. The binding residues in Na2 site are shown in sticks. Bottom panel also includes a schematic summarizing the common architecture among the proteins shown, namely, a conserved site housing a positive charge and formed at the interface of the two helical bundles by TM1 and TM8 helices.