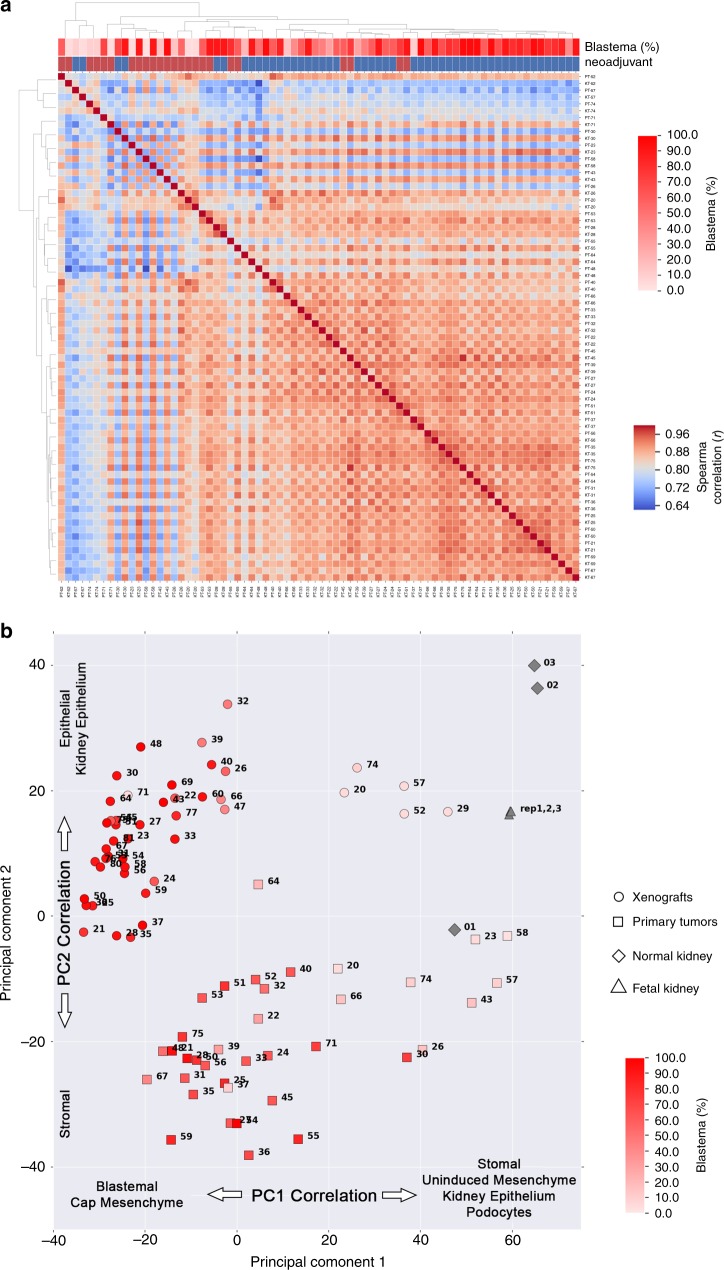
Fig. 5. WTPDX enrich for blastemal gene expression.
a Transcriptomic analysis of RNA-seq data using a Spearman correlation matrix demonstrates highly correlated gene expression between paired WTPDX and primary tumors in most sample pairs. Sample pairs constituting the lowest quartile of Spearman correlation (r < 0.836, blue regions of matrix) were associated with neoadjuvant chemotherapy and blastemal poor primary tumors that often transitioned to blastemal-rich WTPDX (arrows). b Principal component analysis was performed for paired WTPDX (circles) and primary tumors (squares), normal kidney specimens (diamonds), and pooled fetal kidney RNA (triangles) using RNA-seq data. WTPDX clustered distinctly from primary tumors. PC1 explained 21.6% of variance among samples and was inversely correlated with expression of WT blastemal archetype and kidney development cap mesenchyme genes. This increased expression of blastemal genes in WTPDX corresponded to increased percent blastema detected on histologic analysis (heatmap display of percent blastema per specimen shown). PC2 explained 10.5% of variance among samples and was positively correlated with expression of WT epithelial and kidney epithelial genes. KT—xenografts, PT—primary tumor, NK—normal kidney, FK—pooled fetal kidney RNA.