Figure 1.
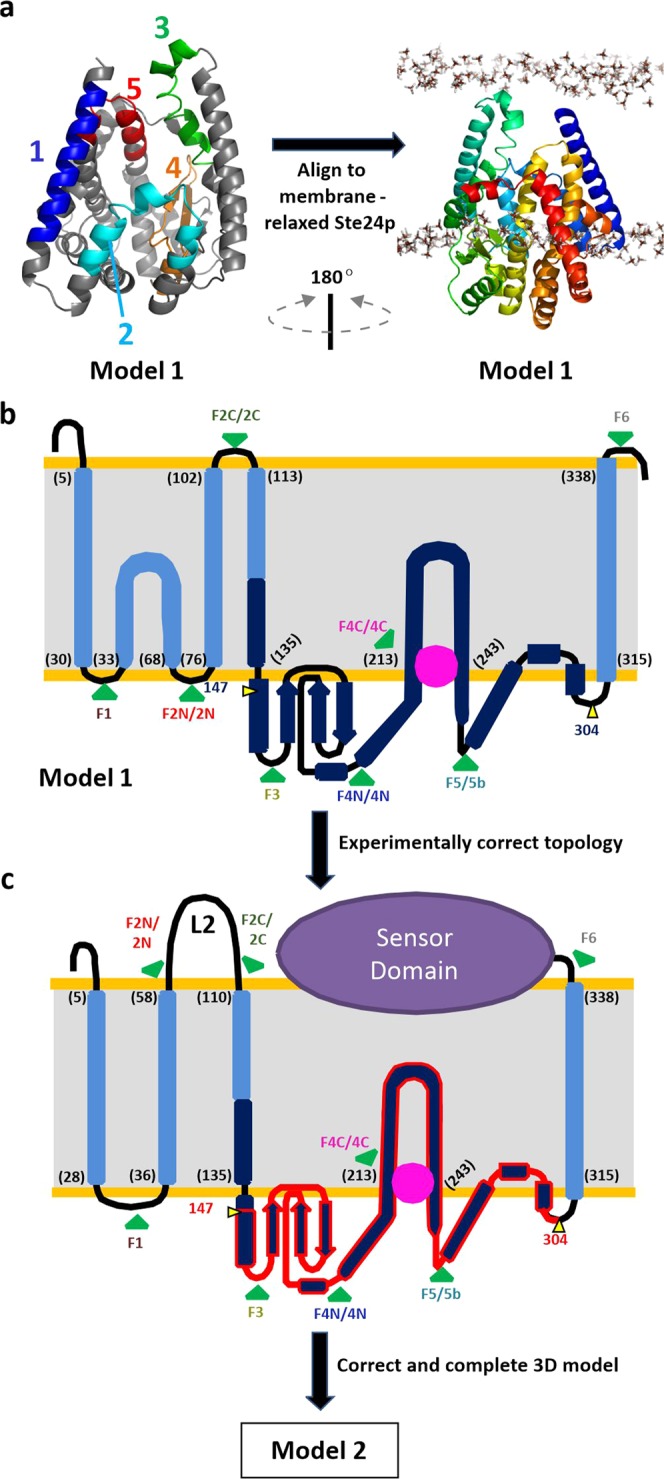
(a) Robetta model (Model 1) of residues 1-340 of the metalloprotease domain of MecR1 (cartoon). The left half maps each of the five transmembrane helices predicted from sequence (Table 1), colored from blue (N-terminal) to red (C-terminal). The right half depicts this model aligned to a membrane-relaxed structure of the Ste24p membrane metalloprotease (see Fig. S2). (b) Flat scheme showing the overall topology of the metalloprotease domain of MecR1 according to Model 1; light and dark blue for the whole homology model, dark blue for the portion backed up by coevolution data. (c) Flat scheme showing the overall topology of full-length MecR1 according to Model 2, which contemplates the results from the experimental mapping of the transmembrane topology. The gluzincin core of MecR1 (MecR1GLZ, residues 147–304) is shown in dark blue with red border. In (b,c), the numbers between brackets indicate the amino acid residue where each membrane-embedded helix is proposed to start or finish; the pink sphere approximates the position of the zinc ion; the green arrows labelled F1, F2N/2N, F2C/2C, F3, F4N/4N, F4C/4C, F5/5b and F6 indicate the positions where either eGFP was fused or a TEV recognition site was inserted for the experimental topology mapping.
