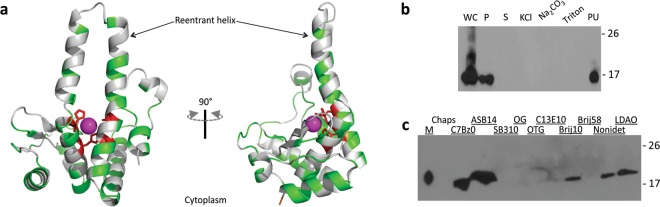
Figure 4.
The gluzincin core of MecR1 is membrane-embedded. (a) The modeled gluzincin domain is shown, with hydrophobic residues displayed in light grey, polar residues in green and the putative metal ligands (H204, H208, E245 and D249) in red. (b) MecR1GLZ.E205A (19.8 kDa) was expressed in E. coli BL21 Star (DE3). IPTG-induced cells (WC, whole cell extract) were sonicated and centrifuged to separate the pellet (P; inclusion bodies and large fragments of membrane) from the fraction containing soluble proteins and membrane vesicles. The latter was subjected to ultracentrifugation to separate soluble proteins (S) from membrane proteins. Lanes labelled KCl, Na2CO3 and Triton correspond to fractions of proteins solubilized from the membrane preparation with 1 M KCl, 0.1 M Na2CO3 and 0.155% Triton X-100, respectively. Lane Pu is the pellet remaining after ultracentrifugation of the membranes treated with Triton X-100. (c) Detergent screening to evaluate conditions to solubilize MecR1GLZ.E205A from membranes. MecR1GLZ was found in the membrane protein fraction (M). The fractions corresponding to solubilized proteins are shown. Both the soluble and insoluble fractions are shown in Fig. S8. MecR1GLZ.E205A could only be solubilized from membranes with the detergents C7Bz0, ASB-14, Brij10, Nonidet and LDAO, being the membrane solubilizing zwitterionic detergent ASB-14 (amidosulfobetaine-14) the one that allowed the highest level of protein recovery. In (b,c), MecR1GLZ.E205A was detected by Western blot using a His-tag specific HRP-conjugated antibody. Numbers on the right indicate the positions of migration of the molecular mass markers (kDa).