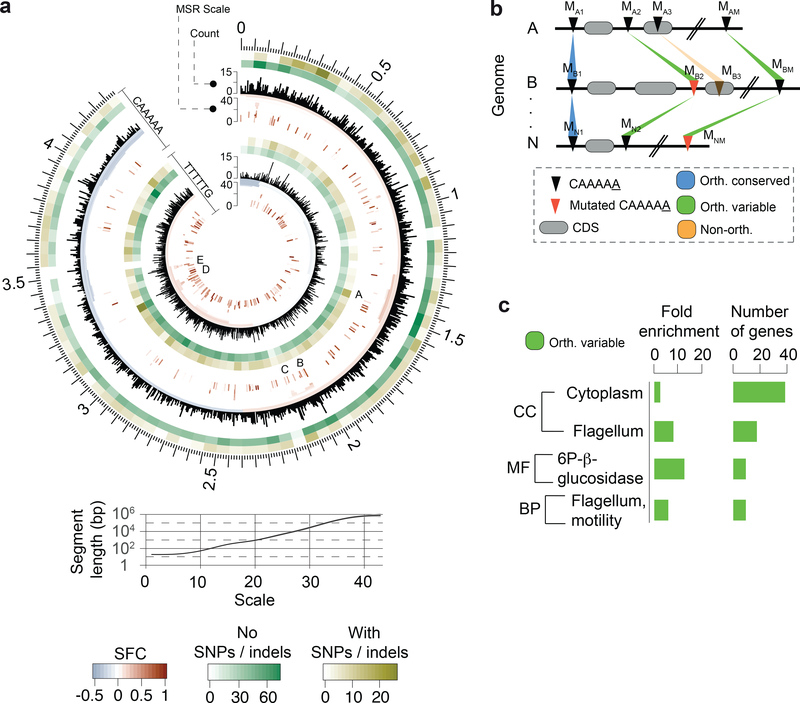
Fig. 3.
Abundance, distribution, and conservation of CAAAAA motif sites. (a) Representation of distribution of CAAAAA sites in both strands of the reference C. difficile 630 genome and corresponding genomic signal obtained by multi-scale signal representation (MSR). Briefly, MSR uses wavelet transformation to examine the chromosome at a succession of increasing length scales by testing for enrichment or depletion of a given genomic signal. While scale values <10 are typically associated with regions <100 bp, genomic regions enriched for CAAAAA sites at scale values >20 correspond to segments larger than 1 kb (i.e., gene and operon scale). Letters (A-E) represent regions with particularly high abundance of CAAAAA motif sites, including genes related to sporulation (e.g., spo0A, spoIIIAA-AH, spoIVB, sigK), membrane transport (PTS and ABC-type systems), transcriptional regulation (e.g., iscR, fur), and coding for multiple cell wall proteins (Supplementary Table 6d). Relation between MSR scale and segment length is also shown. The significant fold-change (SFC) corresponds to the fold-change (log2 ratio) between observed and randomly expected overlap statistically significant at P = 10−6 based on the Z-test. Heatmap layers correspond to the number of orthologous conserved (no SNPs/indels, green-shaded) and orthologous variable (with SNPs/indels) CA5 motif positions. (b) Whole genome alignment of 37 C. difficile genomes (36 isolates + C. difficile 630 as reference) was performed using Mauve. We defined an orthologous occurrence of the CAAAAA motif (black triangles) if an exact match to the motif was present in each of the 37 genomes (conserved, blue-shaded regions), or if at least one motif (and a maximum of n-1, being n the number of genomes) contained positional polymorphisms (maximum of two SNPs or indels per motif) (variable, green-shaded regions). Non-orthologous CAAAAA positions are indicated as orange-shaded regions. The results are shown in Fig. 3a in the form of heatmaps. Numbering in scheme is based on mapping location. (c) DAVID enrichment analysis of genes containing intragenic and regulatory (100 bp upstream the start codon) orthologous variable CAAAAA motif sites. Genes found to over-represent orthologous variable CAAAAA positions include cytoplasm- (e.g., pheA, fdhD, ogt1, spoIVA) and motility-related genes (e.g., fliZ, fliN, fliM, flgL). Single categories were considered significantly enriched at P < 0.05 (one-tailed Fisher’s exact test, FDR corrected) and correspond to 73 out of a total of 617 genes analyzed.