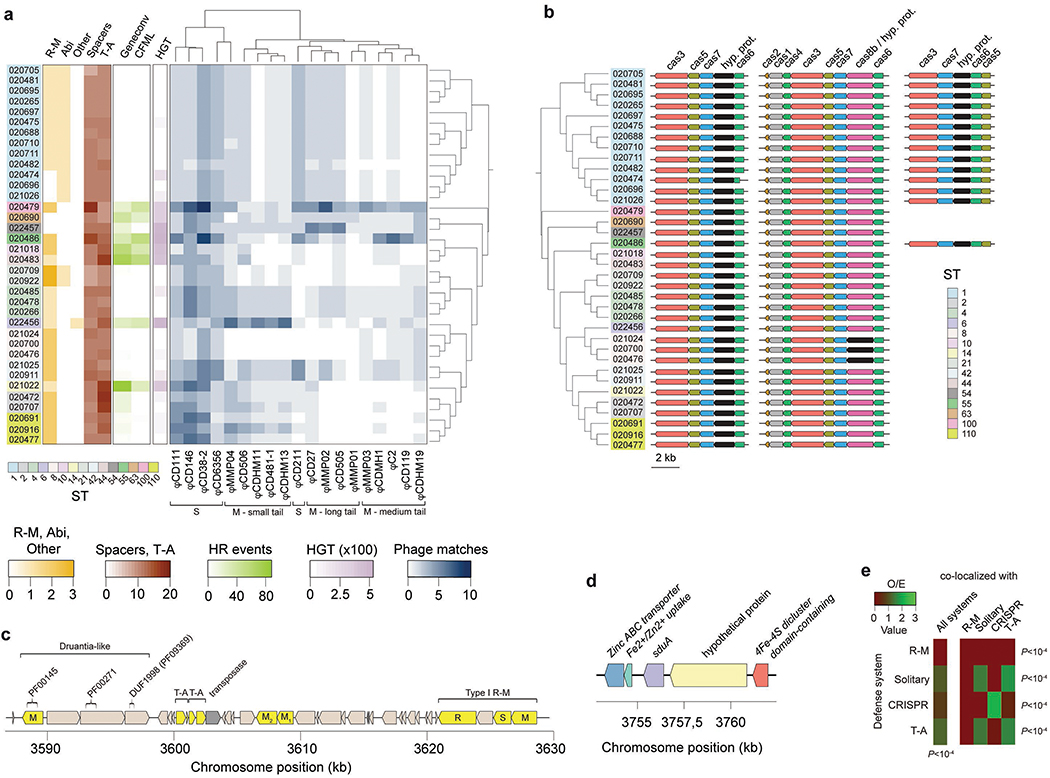
Extended Data Fig. 1. Multiple defense systems and gene flux control in C. difficile.
Multiple defense systems and gene flux control in C. difficile. (a) Heatmap aggregate depicts: abundance of defense systems (R-M, abortive infection (Abi), average number of spacers per CRISPR, toxin-antitoxin (T-A), and Shedu systems (other)), homologous recombination (HR) events (given by Geneconv and ClonalFrameML (CFML)), horizontal gene transfer (HGT, given by Wagner parsimony), and number of phage-targeting CRISPR spacers (Supplementary Notes). Phages were clustered according to their family (Siphoviridae (S), Myoviridae (M)), and tail type. (b) Cas genes detected in C. difficile. Apart from the complete Type-IB gene cluster (cas1-cas8), we also observed two truncated gene clusters lacking cas1, cas2, and cas4. One of the truncated operons was present across all genomes, while the second was restricted to ST-1 and ST-55. (c) Example of a putative ‘defense island’ detected in CD_020472 harboring: a Druantia-like system, two T-A systems, two solitary MTases, and one Type I R-M system. The Druantia-like system is similar to the previously reported Type II Druantia systems62 in the sense that a PF00271 helicase conserved C-terminal domain and DUF1998 (PF09369) are associated with a nearby cytosine methylase. However, it lacks a PF00270 DEAx box helicase. (d) Genomic context of the sduA gene in CD_22456 pertaining to the newly identified Shedu defense system. The gene is located in an integrative conjugative element (ICE) (Supplementary Table 2d). (e) Observed/expected (O/E) ratios for co-localized defense systems (maximum of 10 genes apart). Only the most abundant systems were included in the analysis. Expected values were obtained by multiplying the total number of defense systems by the fraction of co-localized defense systems. P values correspond to the Chi-square test.