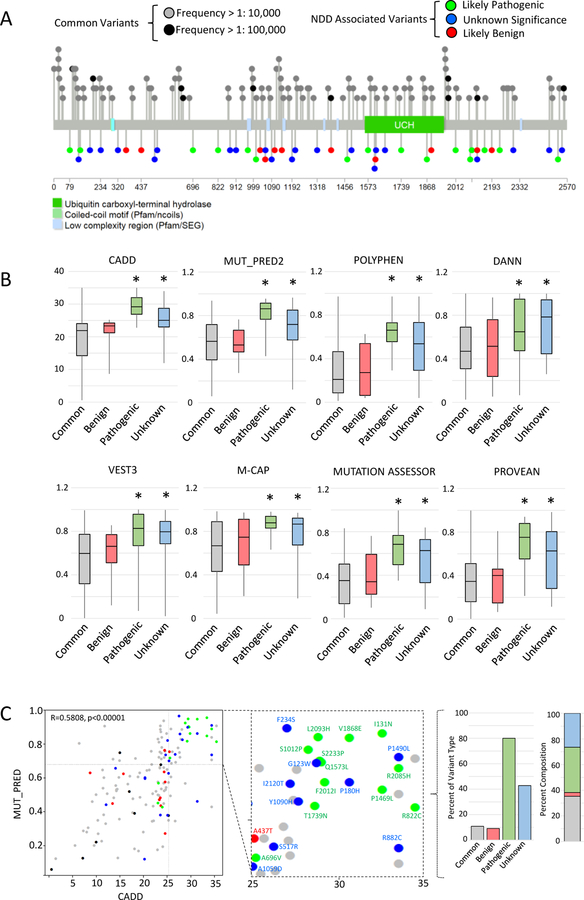
Figure 2. USP9X VUS share in silico signatures with likely pathogenic variants.
A. Protein location of USP9X variants and common variants extracted from gnomAD data base. B. Bulk comparison of common, benign, likely pathogenic and variants of unknown significance by a suite of in silico prediction tools. *significantly different from common variants p<0.05 by Student’s t-test. C. Comparison of CADD and MUT_PRED2 scores reveal clustering of variants of unknown significance with likely pathogenic variants in upper-right quadrant consistent with pathogenicity (CADD >25, MUT_PRED >0.7). Scores are significantly correlated (Pearson’s correlation given). Colour scheme as in A and B. Inset identifies each variant in the ‘pathogenic quadrant’. Graphs show percent of each type of variant, and the overall composition of variant types within the pathogenic quadrant.