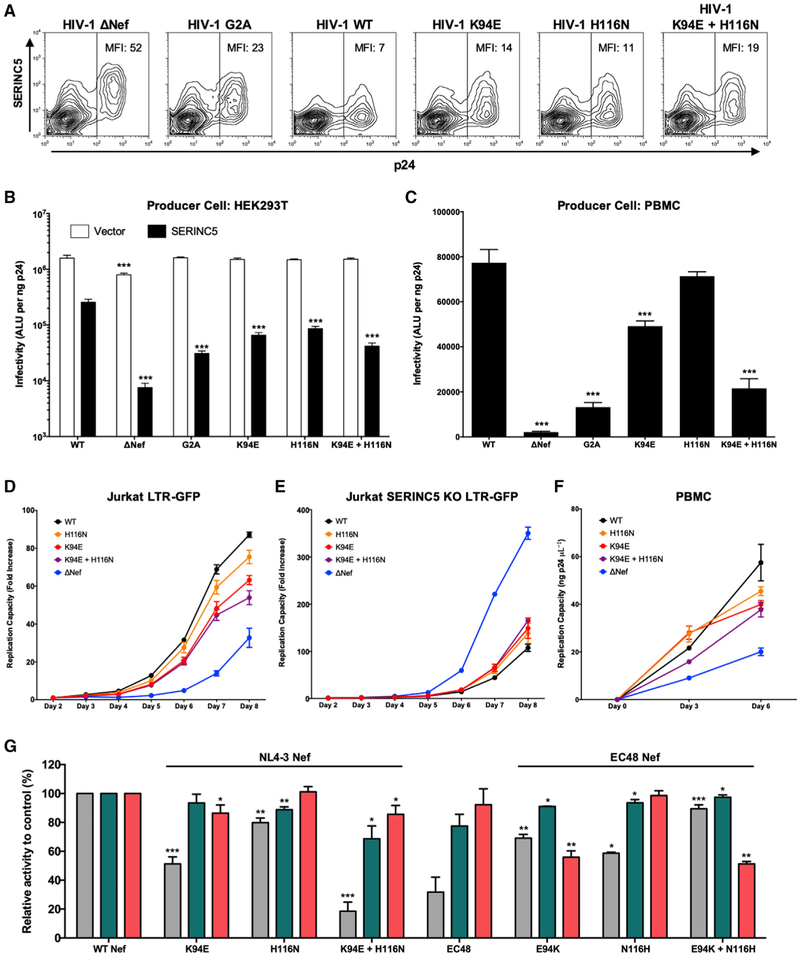
Figure 3. Impact of K94E and H116N Mutants on Nef Function.
(A) Representative flow cytometry plots display SERINC5 surface expression on cells co-transfected with pSERINC5-iHA and pNL4.3. MFI of SERINC5 in the p24+ gate is indicated.
(B and C) Infectivity of NL4.3-derived viruses produced in the absence (white) or presence (black) of SERINC5-iHA (B) or viruses passaged once in healthy donor PBMCs (C) is shown. Results reflect luminescence (absolute light units [ALUs]) following incubation of TZM-bl reporter cells with a normalized amount of virus. Data are representative of two independent experiments (B) or PBMCs from three independent donors (C). Unpaired Student’s t test was used to compare each mutant to NL4.3 (***p < 0.001).
(D and E) Viral replication was examined using Jurkat LTR-GFP R5 cells (D) or those in which SERINC5 was knocked out using CRISPR/Cas9 (Figure S2; STAR Methods) (E). Viral spread was monitored by flow cytometry. The mean (±SD) fold increase in percentage GFP+ cells (from day 2) is reported for each mutant, on the basis of triplicate infections. Results are representative of three experiments.
(F) Replication was also assessed following infection of PHA-stimulated PBMCs. Supernatant was harvested on days 0, 3, and 6 and viral p24 quantified using ELISA. Results are representative of PBMCs from three donors.
(G) The ability of NL4.3 Nef mutants encoding K94E and/or H116N or EC48 Nef reversion mutants encoding E94K and/or N116H to downregulate SERINC5, CD4, and HLA is shown. Bars represent the mean (±SD) function, normalized to control, in at least three experiments. Unpaired Student’s t test was used to compare each mutant with control (*p < 0.05, **p < 0.01, and ***p < 0.001).