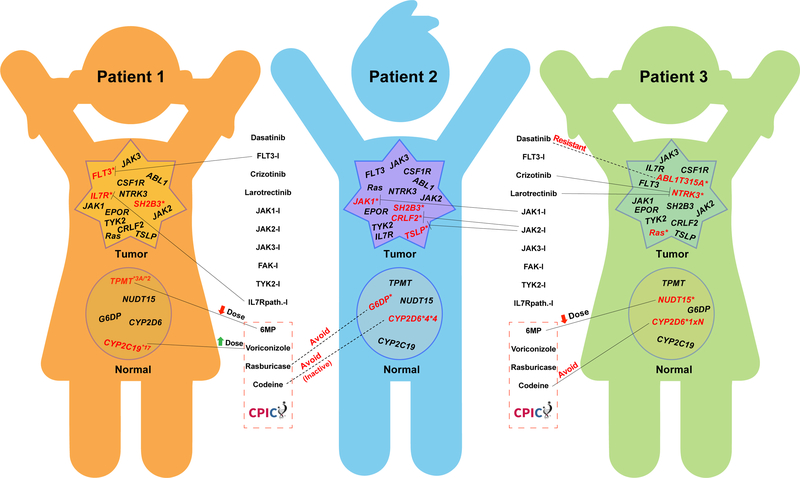
Figure 1. Complexity in selecting optimal medications based on combinations of germline and somatic genome variation in cancer.
Somatic (tumor) and germline (normal) genome variation is reflected for three hypothetical patients with BCR-ABL1-like acute lymphoblastic leukemia (ALL), based on actual genome variants documented from sequencing patients with this disease (17). For each hypothetical patient, genes indicated have already been shown to have functional alterations in ALL (mutations or structural alterations in leukemia cells, inherited variants altering function in germline DNA), and those with mutations in each patient are indicated in red font with an asterisk. Somatic variants are often activating, whereas germline genes are typically loss-of-function (TPMT, NUDT15, CYP2D6) or more rarely gain-of-function (CYP2C19 and CYP2D6 duplication alleles). Potential selection of medications targeting somatic mutations is based on in vitro or in vivo activity of each medication against target proteins. Multiple variants often occur in the same leukemia cell, as documented in prior sequencing studies (17), and often only a subset are treated, as depicted for each patient. All inherited germline variants are essentially always present in the tumor (not depicted). Selection of optimal therapy using inherited germline variants follows evidence-based CPIC guidelines (for medications below the dotted red line). A substantial number of additional somatic and inherited genome variants are known to exist in this disease, adding further complexity to evidenced-based selection of optimal treatment.