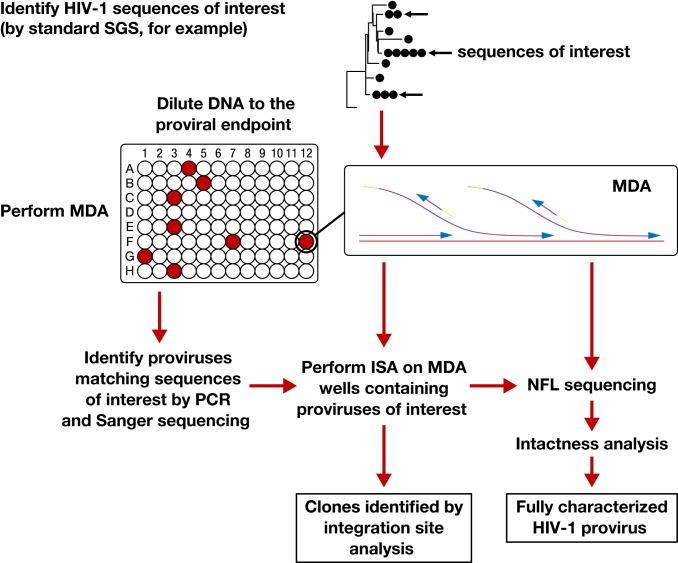
Fig. 1.
Workflow for MDA-SGS. Extracted DNA is aliquoted so that individual proviruses, along with background genomic DNA, can be independently amplified ∼1,000-fold by MDA. This amplification allows the same provirus to be identified by SGS (1–3), analyzed by ISA (integration sites analysis) (1, 10, 22) and NFL (near–full-length) sequencing, followed by further downstream analyses, if needed. Infected cell clones are identified by finding identical integration sites across multiple MDA wells.