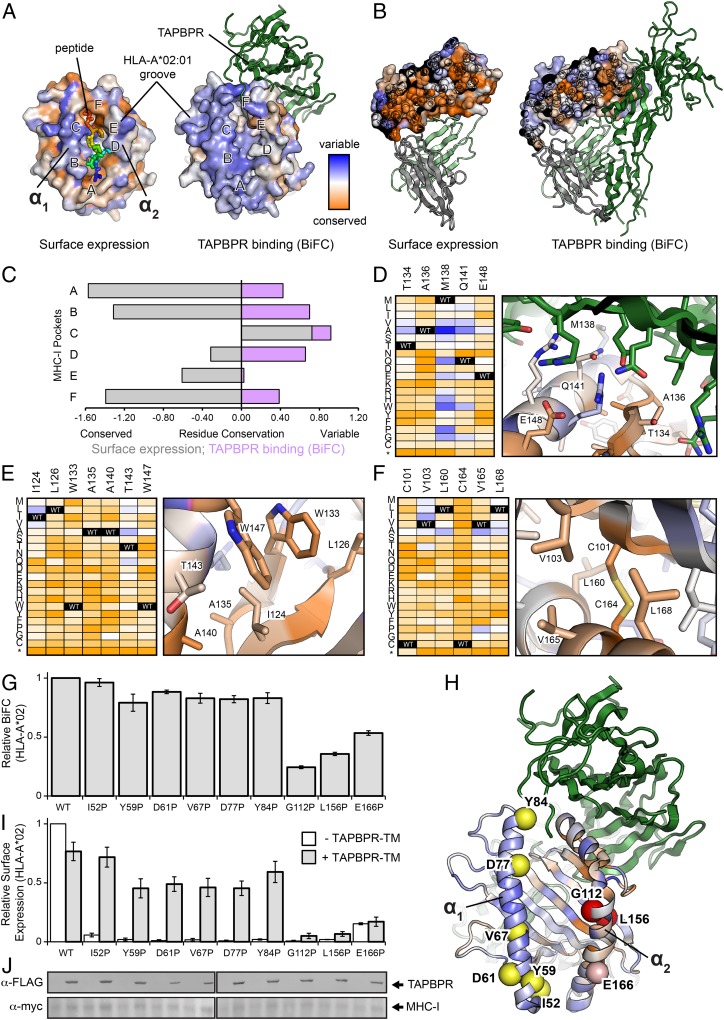
Fig. 1.
TAPBPR recognizes a local conformation of the HLA-A*02:01 α2 domain. (A) Sequence conservation from deep mutagenesis mapped to the surface of HLA-A*02:01. Conserved residues are colored in dark orange, while residues exhibiting mutational tolerance in pale and blue. Residue conservation for HLA-A*02:01 surface expression is shown on the structure of the free pMHC-I molecule (PDB ID code 1HHJ), with bound nonamer peptide colored from blue (residue 1) to red (residue 9). HLA-A*02:01 conservation for TAPBPR (dark green) binding is plotted on the structure of the empty MHC-I, in complex with TAPBPR (PDB ID code 5WER). The location of the A- to F-pockets of the MHC-I groove are noted. (B) Cross sections through the core of the α2 domain, colored by conservation as in A. TAPBPR is dark green, β2m is pale green, and the α3 domain is gray. (C) Conservation and variation of residues in the MHC-I pockets for surface expression (gray) or TAPBPR binding by BiFC (purple). (D–F) Heat maps of mutation log2 enrichment ratios for HLA-A*02:01/TAPBPR BiFC (depleted mutations are orange, enriched mutations are dark blue) shown alongside modeled structures of the (D) α2 domain/TAPBPR interface, (E) the hydrophobic core between the α2–1 helix and β-sheet, and (F) the core between the α2–2 helix and β-sheet. (G) Relative BiFC signal between TAPBPR-TM-VC and MHC-I–VN for different proline substitutions in either the α1 or α2 helices, or β-sheet underlying the α2 helix. (H) Location of proline substitutions on the MHC-I groove mapped onto the TAPBPR complex structure (PDB ID code 5WER). (I) Relative surface expression for the different MHC-I proline substitutions in the presence or absence of TAPBPR-TM. (J) Immunoblots comparing total expression levels for TAPBPR-TM (α-FLAG) and HLA-A*02:01 (α-myc) constructs. Lanes are aligned with graphs above.