Graphical abstract
Abbreviations: BAC, Bacterial Artificial Chromosome; bp, base pair; ddNTP, 2,3-dideoxynucleoside triphosphate; dNTPs, deoxynucleoside triphosphates; HapMap, haplotype map; HGP, Human Genome Project; HMW, high molecular weight; NGS, Next Generation Sequencing; OLC, Overlap-Layout-Consensus; QV, Quality Value (QV); SBS, Sequencing by Synthesis; SMRT, Single Molecule Real-Time; SNPs, Single Nucleotide Polymorphisms; SRA, Short Read Archive; SV, Structural Variant; TGS, Third Generation Sequencing; WGS, Whole Genome Sequencing; ZMW, Zero-Mode Waveguide
Keywords: Genome assembly, Sequencing, Reference, Bioinformatics, Next-generation, Third-generation
Abstract
Genomes represent the starting point of genetic studies. Since the discovery of DNA structure, scientists have devoted great efforts to determine their sequence in an exact way. In this review we provide a comprehensive historical background of the improvements in DNA sequencing technologies that have accompanied the major milestones in genome sequencing and assembly, ranging from early sequencing methods to Next-Generation Sequencing platforms. We then focus on the advantages and challenges of the current technologies and approaches, collectively known as Third Generation Sequencing. As these technical advancements have been accompanied by progress in analytical methods, we also review the bioinformatic tools currently employed in de novo genome assembly, as well as some applications of Third Generation Sequencing technologies and high-quality reference genomes.
1. Introduction
A genome is the complete genetic information of an organism or a cell. Single or double stranded nucleic acids store this information in a linear or in a circular sequence. To precisely determine this sequence, progressively more efficient technologies characterized by increased accuracy, throughput and sequencing speed have been developed. Nonetheless, sequencers can generate sequences, known as reads, comprised only in defined ranges of lengths, usually far shorter than the size of the genomes investigated. The complete genome sequence has to be deduced from the overlaps of these shorter fragments, a process defined as de novo genome assembly. Historically, mostly due to time and cost constraints, only an individual per species was addressed, and its sequence generally represents the ‘reference’ genome for the species. These reference genomes can guide resequencing efforts in the same species, acting as a template for read mapping. They can be annotated to understand gene function or used to design gene manipulation experiments. Sequences from different species can be aligned and compared to study molecular evolution. Due to the impact of reference genomes in all these downstream applications, it is paramount that their sequence is as much complete and error-free as possible. In the last 50 years of the XX century, available sequencing technologies allowed to focus mostly on relatively small genomes. Since the new millennium, novel platforms, known as Next Generation Sequencing (NGS), have been developed to address larger genomes, in a process called Whole Genome Sequencing (WGS). In the two decades following the advent of WGS, NGS has become increasingly more efficient and affordable. In parallel, new sequencing technologies have emerged that promise to revolutionize the field and generate genomes of higher quality.
2. Historical background
2.1. Sequencing nucleic acids in the XX century
In 1953 Watson and Crick published their seminal paper unraveling the double helix structure of DNA [1]. In their work they noted “so far as is known the sequence of bases along the chain is irregular” and “the sequence of bases on a single chain does not appear restricted in any way”, two features that entail a role of DNA in the storage of genetic information, and highlight the importance of determining the exact sequence of bases along the chain. Year 1953 also marked the first sequencing of a biological molecule. Thanks to a refined partition chromatography method, Sanger was able to sequence the two chains of insulin protein [2], [3]. In his approach, proteins were randomly fragmented, the fragments were individually read, and sequences from each fragment were overlapped to yield a complete, consensus sequence. Proteins were sequenced before nucleic acids, but many principles remained the same, paving the way to modern DNA sequencing. When in 1965 it came to nucleic acids, first it was the turn of the 76 bases long alanine tRNA from Saccharomyces cerevisiae, as methods to cleave RNA fragments were available since the 1940s [4]. As for the insulin, RNA was first fragmented with RNAse A and RNAse T1, the pieces were separated by chromatography, and the degradation products were used to deduce its sequence. At the end of the 1960s, RNA sequencing was still ahead of DNA sequencing. In 1968, the 12 bases long complementary extremities of phage λ cos-site were the first DNA molecules sequenced [5]. The same year, a team including Sanger determined the 120 base pair (bp) long 5s rRNA using 32P-labeled RNA and a paper fractionation-based approach [6]. In 1972, Fiers sequenced the first gene, the 510 bp of the coat protein gene from the RNA virus phage MS2 [7]. In 1973, Gilbert and Maxam reported the 24 bases of Escherichia coli lactose-repressor binding site by copying its DNA into RNA [8]. In 1976, phage MS2 was the first organism to have its 3,569 bp RNA genome completely sequenced [9] (Fig. 1). In the mid 1970s, DNA sequencing was about to flourish. In 1975, Sanger and Coulson developed the ‘plus and minus’ method for DNA sequencing and used it to determine two short regions in phage φX174 single-stranded DNA [10]. In this method, four reaction mixtures are prepared each containing the template DNA, a primer, DNA polymerase and the four deoxynucleotides, one of which is radiolabeled. These reactions generate a population of newly synthesized DNA strands of different lengths. Reactions are then split into four pairs of “plus” and “minus” mixtures: the minus mixtures contain three deoxynucleoside triphosphates (dNTPs) and, as a result, the DNA strand is extended up to the missing nucleotide. In the “plus” reactions only one nucleotide is added to each of the four aliquots and T4 DNA polymerase is used. The exonuclease activity of the T4 DNA polymerase is harnessed to degrade DNA from 3’-end up to the nucleotide that is supplied in the reaction. The products of the eight reactions are loaded on polyacrylamide gels [11]. X-ray films are used to produce carbon-copy images of the gel slabs and migration distance is employed to determine the nucleotide order. In 1977, using this approach Sanger determined the 5368 bp genome of phage φX174, the first DNA genome to be sequenced [12] (Fig. 1). The same year, he developed a new method that could decode fragments of approximately four hundred bases in a day [13]. The ‘Sanger’ method relies on four separate polymerization reactions performed using tritium-radiolabeled primers, where each reaction is supplied with small amounts of one chain-terminating 2,3-dideoxynucleoside triphosphate (ddNTP) to produce fragments of different lengths [13]. When the DNA polymerase incorporates a ddNTP at the 3’-end of the growing DNA strand, it lacks a 3’-hydroxyl group and chain elongation is terminated [14]. Similarly to the ‘plus and minus’ approach, the sequence is deduced by comparing the size of the fragments. The same year, Maxam and Gilbert proposed an alternative, purely chemical, approach [15]. While extremely popular for years, with the improvement of the chain-termination method, this approach fell out of favor due to its technical complexity, use of hazardous chemicals, and difficulties to be scaled up.
Fig. 1.
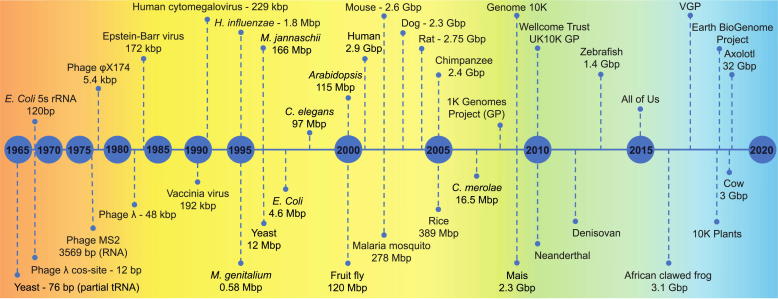
Milestones in genome assembly. Timeline illustrating many of the major genome assembly achievements ranging from the beginning of the sequencing era to the large-scale genome projects currently ongoing. Each genome or genome project (GP) is placed under a color-coded background according to the sequencing approach adopted. Light red: early sequencing methods, Yellow: Sanger-based shotgun sequencing, Green: NGS, Light blue: TGS. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. Shotgun sequencing
To hasten the sequencing process, in 1979 Staden proposed the idea of ‘shotgun sequencing’, where bacterial vectors are used to clone random fragments of a long DNA molecule. Fragments are then sequenced in parallel and reads are assembled using their overlaps [16], allowing to sequence larger genomes in shorter times. In 1981, Messing developed the first shotgun sequencing protocol using the single-stranded M13 phage vector [17]. Only one year later, Sanger employed this method to assemble the entire 48,502 bp-long genome of phage λ [18]. In 1984, the genome of Epstein–Barr virus B95-8 strain, comprising 172,282 bp, was determined using the same approach [19]. Starting from the 1980s, many full-genome sequencing projects were launched and succeeded (Fig. 1), as testified by the increasing availability of data in dedicated repositories [20]. GenBank, the US National Institute of Health (NIH) sequence database, was founded in 1982 with about half a million bases, and by the end of the decade it included over 40 million bases [20]. This growth rate has never ceased, with almost 10-fold increases every 5 years. “Sequences, Sequences, and Sequences” was the title of a Sanger’s article in 1988, suggestive of the general excitement [21]. In the 1980s and 1990s, technological breakthroughs combined with the adoption of industrial processes increased throughput and reduced sequencing errors (Box 1.1). These advancements were further accelerated by the Human Genome Project (HGP), which aimed to produce genetic maps, physical maps, and finally the complete sequence of human chromosomes (Box 2). Moreover, the competition with Celera, a private company that also decided to assemble the human genome, stimulated both initiatives to obtain results rapidly [22], [23] (Box 2). With the HGP, companies realized that sequencing could be a profitable business and the market competition gave birth to a plethora of sequencing technologies, collectively referred to as Next-Generation Sequencing (NGS) (Box 1.2).
Box 1. The industry of DNA sequencing.
Box 1.1. Automation of Sanger Sequencing
In the early 1980s, DNA sequencing was performed with the original Sanger method, involving four sequencing reactions in four separate tubes using radiolabeled primers. In 1985, Smith and Hood synthesized fluorescent DNA primers [117] and used them for the partial automation of Sanger sequencing [118]. In this method, known as dye primer sequencing, four dyes distinguished by their fluorescent emission spectra are covalently attached to the oligonucleotide primers and employed in distinct dideoxy extension reactions. The products are then run together on a polyacrylamide gel and Fluorescent Energy Resonance Transfer is used to read the sequence. In 1987, Prober described dye terminator sequencing, a novel approach based on four chain-terminating dideoxynucleotides, each carrying a succinyl fluorescein dye with a different emission spectra [119]. The distinct fluorescence signals allowed reactions to be performed in a single tube. In 1986, Applied Biosystems, a company founded in 1981 focused mostly on protein sequencing, switched to nucleic acids and released the first commercially available four-color fluorescence automated DNA sequencer (ABI 370A) based on these advancements. A scanning system allowed multiple samples to be run simultaneously, with automatic computer-based base calling. The machine was able to handle 32 samples per run, quadrupling the throughput. The next year this approach was refined by introducing an optimized T7 DNA polymerase [120]. This enzyme was highly processive, lacked proofreading 3′ to 5′ exonuclease activity and efficiently used nucleotide analogs, allowing to generate around 1000 bases per day [121]. Two years later, the introduction of the thermostable Taq DNA polymerase and of cycle sequencing greatly reduced template requirements and facilitated miniaturization. Now the sequencing reaction could be performed at 72 °C and repeated multiple times in the same tube, achieving linear amplification of the template [122]. The technological advancements of the 1980s were further reinforced and extended in the 1990s. Major milestones included the introduction of fluorescent boron-dipyrromethene dyes (bodipy) more effective than conventional dyes [123], magnetic bead-based DNA purification methods to simplify the automation of pre-sequencing steps [124], and capillary electrophoresis, which eliminated the pouring and loading of gels, while also simplifying the identification and interpretation of the fluorescent signal [125]. These rapid advancements were further prompted by the HGP race (Box 2). In 1995, Applied Biosystems replaced ABI 370A with another slab-gel electrophoresis system, ABI Prism 377, which could deal with up to 96 lanes at a time. One year later was released ABI Prism 310, the first capillary DNA sequencer, and in 1998 the high-throughput ABI Prism 3700. In 2001, it was the turn of the 16-capillary ABI Prism 3100, and in 2002 that of ABI 3730xl provided with 48 to 96 capillaries, where sequences included QVs. By 1998, routine DNA sequencing of 1000 bases was achieved in less than one hour [126]. During the great period of excitement of the 80s and 90s, even a completely new method for DNA sequencing by stepwise dNTP incorporation was developed [127], [128]. Known as “pyrosequencing”, this approach relies on measuring the enzymatic luminometric signal generated by pyrophosphate release during DNA polymerization [129]. This strategy has several advantages over Sanger’s approach, including the use of natural nucleotides and the possibility of observing nucleotide synthesis in real time [130].
Box 1.2. A $1000 genome
In the 1990s, Applied Biosystems was the undisputed market leader. In contrast with this earlier monopoly, in the 2000s many companies started to offer different technologies with progressively higher sequencing throughput at lower costs. These included 454, Solexa, Illumina, Agencourt, Complete Genomics, Applied Biosystems and Ion Torrent. Following the enthusiasm generated by the HGP (Box 2), the ultimate goal became making the cost of sequencing a whole human genome more affordable, achieving a price below $1000 per genome. The first generation of NGS sequencers was based on pyrosequencing and initially produced by Pyrosequencing AB. Through a series of acquisition, this technology was later commercialized by 454 Life Sciences, which in 2003 introduced on the market the first NGS DNA sequencer, the GS20. The GS20 used single-molecule template synthesis of small, bead-bound DNA fragments in a water-in-oil emulsion clonal PCR (emPCR) [131], and dNTP incorporation detection by CCD sensors beneath the surface of about one million microwells [132]. The system produced approximately 400–500 bp-long reads, had 99% accuracy and could sequence up to 25 million bp in a single 4-hour run at less than one-sixth the cost of conventional methods. In 2003, Solexa started to develop a new sequencing approach, later known as Sequencing by Synthesis (SBS) [133]. Next year Solexa acquired from Manteia the colony sequencing technology, also known as bridge amplification [134]. In bridge amplification, tightly clustered copies of individual molecules, or “polonies” [135], are produced on a surface from an immobilized template library [136]. The resulting stronger signal enhanced the accuracy of base calling, while reducing the cost of the system optics. In 2005, Solexa added to its method an engineered DNA polymerase [137] and the recently developed reversible terminators that can be washed away, allowing for continuous extension steps [138], [139]. The resulting platform was able to image and determine each single nucleotide added to all the DNA fragments placed on the surface of a flow cell. Genome Analyzer, the first Solexa commercial sequencer, was launched in 2006. In contrast to GS20, Solexa sequencer had a higher throughput of 1 Gbp in a single run, but sequencing reads were shorter, only 35 bp long [140]. However, a great advantage was that these represented paired end reads (i.e. both DNA strands were simultaneously sequenced), allowing to size the gap between relatively distant sequences in a DNA fragment. In 2007, Solexa was acquired by Illumina, a company founded in 1998 that by 2001 had started offering SNP genotyping services. The 2007 was also the year of SOLiD from Applied Biosystems. This technology was based on a ligation strategy, relying on the specificity of DNA ligases to ligate fluorescent oligonucleotides to templates in a sequence-dependent manner [141]. This method was later reported to have some issues in sequencing palindromic sequences [142] and was consequently abandoned. A new approach based on proton detection in semiconductors was released in 2011 by Ion Torrent [143]. This approach generated 100 bp long reads by measuring the pH variations induced by hydrogen ions released during nucleotide addition in DNA synthesis, using a semiconductor sensor. Parallel measurements of multiple templates were carried out in microwell plates where each of the four nucleotides was added in succession. The process of reading each nucleotide could occur in a few seconds, without imaging and therefore at a considerably lower cost. Based on an advancement of this technology, in 2012 Ion Torrent released a more powerful machine called Ion Proton, which the company claimed could have allowed to sequence a human genome in a single day for the price of $1000. However, similarly to SOLiD, Ion Torrent platforms often mismeasured the length of homopolymers [144] and were therefore abandoned. Due to the limitations of the other sequencing platforms, the SBS approach commercialized by Illumina ultimately prevailed. By 2014, the company appeared to have reached a position of near monopoly [145], holding 70% of the market for DNA sequencers and accounting for more than 90% of all DNA data produced [146]. The same year, the company announced the HiSeq X Ten, claiming that forty of these machines would have been able to sequence more genomes in one year than had been produced by all other sequencers to date, allowing large-scale WGS for $1000 per genome [147]. This goal has been exceeded in 2017, when Illumina released NovaSeq, a sequencer that can output over 3000 Gbp in a single run. However, these short read-based genomes are partially incomplete and represent only draft assemblies, that can be improved through complementary sequencing and scaffolding approaches (Section 3). Consequently, the new frontier is becoming to make hybrid genome assembly also more affordable.
Box 2. The reference human genome.
In 1988, the US Department of Energy (DOE) and NIH received funding and formalized an agreement to “coordinate research and technical activities related to the human genome” [148]. Watson was appointed to lead the NIH component and the official kickoff was in 1990 with the publication of a joint research plan for the next five years. The deadline for project completion was set to 2005, however Watson resigned in 1992 due to the persistent confrontations with the NIH Director [149]. The following year Collins was appointed in his place and traced a new five-year plan for the HGP [150]. By 1994 the HGP team published the first linkage map of the human genome [151]. Despite the successes with smaller genomes, in 1998, formally halfway through the schedule for completing the HGP, only 50 Mb of human sequence had been determined [152]. The HGP team had developed an accurate but laborious strategy named “hierarchical shotgun” sequencing. The pipeline involved cloning large fragments of human genome into Bacterial Artificial Chromosomes (BACs). BACs were then fragmented, size-selected and subcloned. DNA from individual clones was used as a template for automated Sanger sequencing [22]. Nonetheless, HGP leaders announced the intent to complete the project in 2003, two years ahead of previous projections [153]. However, a private effort led by Venter’s company Celera decided to rival the public HGP, accelerating the events. Counter to HGP leaders, the team at Celera believed direct shotgun sequencing without BAC intermediates to be the fastest and most effective approach also to sequence larger genomes [154]. Building upon the experience gained at The Institute for Genomic Research (TIGR) with bacteria, Celera team rapidly sequenced the 175 Mbp of Drosophila melanogaster genome using an internally-developed WGS strategy and a new genome assembler based on a more effective OLC approach [155] (Box3). The success with D. melanogaster prompted Venter to turn to the human genome. In March 1999, HGP leaders announced the successful completion of the pilot phase and at the beginning of 2000 the HGP team deposited two of the three Gbp of the human genome [156]. Progress was fast also at Celera, and in less than two years they generated their draft of the human genome. The race culminated in June 2000 with a joint announcement. The HGP officially released the sequence in February 2001 [22] and Celera published its genome assembly one day later [23]. At that time, both the HGP and Celera teams had published only working drafts of the human genome. The most complete assembly, from the HGP, represented about 90% of the human genome, with only 25% in curated form [22]. Due to the direct shotgun approach employed, the assembly generated by Celera was even less accurate [157]. A high-quality reference would have required three more years of work by the HGP. Finally, in October 2004 it was published the near-complete human genome sequence [158]. Until today, the human reference genome has been continuously improved by the Genome Reference Consortium [159], resulting in numerous releases. The quest for higher quality human reference genome(s) is still on-going as indicated by the open calls for proposals aimed to produce more accurate assemblies, as well as to generate a ‘pan-genome’ that can capture nearly all human variation. In parallel, also the affordability of sequencing entire genomes has been promoted. The cost of the HGP was $2.7 billion in fiscal year 1991 dollars [160]. Thanks to the development of progressively refined sequencers based on Sanger sequencing, per base cost dropped over 100-fold during the HGP [157]. Moreover, after completion of the HGP, the NHGRI started the “Revolutionary DNA Sequencing Technologies program” awarding millions of dollars in research grants to further support the development of new sequencing approaches, and paving the way to more affordable genomes (Box 1.1 and 1.2).
2.3. DNA sequencing in the third millennium
From a technical standpoint, the new generation of sequencing approaches resulted from a combination of advancements in microfabrication, high-resolution imaging and computational power [24] (Box 1.2). All NGS approaches rely on a ‘library’ preparation using native or amplified DNA. In a classical protocol, after DNA fragmentation and fragment size selection, adapters are ligated to the ends of each fragment. This is generally followed by a step of DNA amplification. The resulting library is loaded on a flow cell and sequenced in Massive Parallel Sequencing reactions [25]. Each reaction usually involves the stepwise, polymerase-mediated incorporation of fluorescently labeled deoxynucleotides in an elongating DNA chain immobilized on a surface (Box 1.2). Results from the blossom of high-throughput sequencing technologies have been innumerable. The power of the new methods was proved by the cost-effective and rapid resequencing of many milestone genomes. During this period of excitement, genome drafts for most eukaryotic model species were produced (Fig. 1). NGS also allowed extremely novel applications, including whole-exome sequencing [26], high-throughput RNA sequencing (RNAseq) [27], chromatin immunoprecipitation followed by sequencing (ChIP–seq) [28], and genome-wide epigenetic landscape determination [29]. Human genotyping became popular, with millions of Single Nucleotide Polymorphisms (SNPs) from hundreds of individuals identified by projects as the International HapMap Project [30]. Human genome resequencing started to become more affordable. The first individuals to have their genome fully re-sequenced were Venter in 2007 [31] and Watson in 2008 [32]. In 2011, six-year-old Nicholas Volker was reported as the first patient saved by DNA sequencing, as his one in 1 billion genetic mutation of XIAP gene resulted to be treatable with cord transplant [33]. Efforts in human genome sequencing were resumed by the 1000 Genomes Project (2008–15) [34], and more large-scale human genome projects have constantly been and are being proposed such as the Wellcome Trust UK10K in 2010 and the All of Us in 2015 [35] (Fig. 1).
3. Genome sequencing and assembly in the Third Generation Sequencing era
Starting from the 2010s, radically new technologies opened the era of Third-Generation Sequencing (TGS). TGS definition may vary, but it is generally given to technologies capable of sequencing single DNA molecules without amplification. Nowadays, these technologies allow to produce reads far longer than NGS, each spanning several to hundreds kbps. In the context of genome assembly, the availability of long reads constitutes a great advantage, as the difficulty of detecting overlaps between NGS short reads reduces the ability of generating long continuous consensus sequences [36], impacting the overall quality of the assemblies [37], [38], [39]. This issue is further exacerbated by the presence of repeated and low complexity DNA regions [40]. As more complex genomes are addressed, the impact of repeat elements increases [41]. In these cases, the final assembly continuity may be reduced even adopting refined NGS-based approaches, such as 10X Genomics linked reads, where barcodes are associated with individual high molecular weight (HMW) DNA molecules prior to NGS library preparation and later used to group short reads sharing the same barcode as originating from the same HMW DNA fragment [42], [43]. Extra-large genome sizes such as those of amphibians [44], where low-complexity regions account for most of the ‘extra’ DNA [45], constitute a practically insurmountable barrier for NGS-based WGS (Fig. 1). TGS enables to address these shortcomings to generate genome assemblies of unprecedented quality [46], [47], [48] (Fig. 1).
3.1. Single-molecule sequencing
The first single-molecule sequencing technology, based on fluorescence detection and Sequencing by Synthesis (SBS), was developed by Quake and commercialized in 2009 by Helicos BioSciences [49], [50]. It worked similar to Illumina sequencers, but without any bridge amplification, thereby avoiding DNA amplification-associated biases. However, it was slow, expensive and produced relatively short reads, around 35 bp long. Two single-molecule approaches were developed in the last decade that overcame these disadvantages. The first approach, Single Molecule Real-Time (SMRT) sequencing was developed by Craighead, Korlach, Turner and Webb and was further refined and commercialized by Pacific Biosciences (PacBio) since 2011. The second approach, Nanopore sequencing, was first hypothesized in the 1990s, but developed into a technology only decades later [51] and commercialized by Oxford Nanopore Technologies (ONT) since 2005. SMRT sequencing normally generates reads with average median length of approximately 10–20 kbp, but also several reads longer than 50 kbp. During library preparation, specific hairpin adapters are ligated to both ends of double-stranded DNA molecules to circularize them, forming a structure known as SMRTbell [52]. SMRTbell libraries are then loaded onto a SMRT cell containing hundreds of thousands Zero-Mode Waveguide (ZMW) nanowell arrays [53]. Each ZMW contains a polymerase immobilized at the bottom that can bind a primer complementary to the hairpin adapters of the SMRTbell and start the replication process [54]. Individual sequencing reactions take place within each ZMW, corresponding to the smallest available volume for light detection (20 zeptoliters each), and consist in the incorporation of the four different fluorescent-labeled nucleotides in succession using the SMRTbell DNA as complementary template. Each incorporation event produces a specific emission signal that is recorded for real-time base-identification by an ultrahigh resolution camera coupled with a computer [54]. A ‘movie’ of light pulses, corresponding to a sequence of bases, is named Continuous Long Read (CLR). The individual base-calling accuracy of these reads is far less accurate than Illumina short reads, with ~1 error every 10 nucleotides [55] compared to the reported ~1 error every 1000 nucleotides using NGS [56]. However, in contrast to NGS, there appears to be little or no sequencing bias [57] and error profiles seem to be purely random, allowing high-quality in the final consensus sequence [58]. Interestingly, the circular nature of DNA molecules in SMRTbells allows to read the same sequence multiple times. Over the years, PacBio has further developed this concept to increase base-calling accuracy, culminating in 2019 with the release of a new method named HiFi (‘High Fidelity’) long-reads, which can generate Circular Consensus Sequences (CCSs) approximately 10–20 kbp-long, potentially as accurate as Illumina short reads [59]. The advantages of SMRT sequencing over NGS have come at the price of higher per base sequencing costs. In 2019 PacBio has released Sequel II, a revised platform based on new chemicals and equipped with a novel 8 million ZMW nanowells SMRT cell, theoretically able to generate a throughput of 160 Gb per SMRT Cell, with a concomitant drop of up to 8-fold in sequencing costs.
Nowadays, the direct competitor of SMRT sequencing for both quality and price is Nanopore sequencing. Nanopore sequencing uses genetically modified bacterial nanopores inserted into an artificial lipid bilayer, placed in individual microwells tens of micrometers wide and arrayed on a sensor chip [60], [61], [62]. As each single DNA strand travels through a channel, it disrupts the current running through the pore, and the change is measured by a semiconductor sensor. Since each base disrupts the electric field in a slightly different way, the recorded current changes can be translated into a DNA sequence. The final result is similar to that of SMRT sequencing, with long reads of low base-calling quality. Read length is in the order of 10–20 kbp, and the longest molecule sequenced to date was 2.3 Mbp long [63]. This places ONT reads as the longest DNA reads generated so far, allowing to sequence through repeats where even PacBio reads may fail. Furthermore, since ONT technology relies on the detection of an electrical rather than an optical signal, Nanopore devices can be as small as a USB stick. In 2016, such portability allowed to sequence Ebola virus at field sites in West Africa in less than 60 min [64]. However, ONT reads appeared to have sequencing biases difficult to be corrected [65]. In 2019, Nanopore has released into early access R10, a new nanopore with a double sensor for improved base calling [66].
3.2. Advantages of Third Generation Sequencing over Next Generation Sequencing
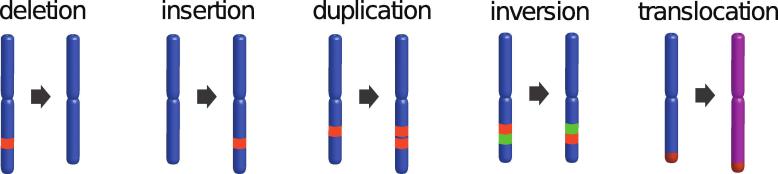
TGS has been shown to considerably improve the quality of genome assemblies [58], [67]. Ultralong read lengths and low sequencing-context bias (GC-content or low-complexity) enable to assess many regions missing from NGS-based assemblies [39], [68], providing a more uniform coverage along the genome and increased structural accuracy of the resulting assemblies. Moreover, in diploid (or polyploid) genomes TGS allows the generation of long phased blocks of haplotypes (haplotigs), where the paternal and maternal contributions to a homologous region of the chromosome are reported separately [69]. Avoiding chimeric genomes, where paternal and maternal haplotypes are mixed or even collapsed, facilitates accurate mapping of sequencing reads for structural variants (SV) detection, including length variations in highly repeated motifs, other large insertion and deletion events (indels), duplications, inversions and translocations [70], [71] (Fig. 2). Indeed, while SNPs were long regarded as the most relevant type of genetic variation, it is now clear that SVs (>50 bp) also play a key biological role [72], [73]. SVs comprise a large portion of bp accounting for interspecies genotypic differences [74], implying that an important fraction of genomic variation has so far escaped detection by NGS. This could explain much of the “missing heritability” problem [75]. Accordingly, evidence for the importance of SVs in determining both simple and complex phenotypic traits is constantly growing [76], [77]. Finally, another advantage of TGS over NGS is that it provides the simultaneous capability of characterizing a variety of epigenetic marks along with DNA sequencing [78], [79]. This is achieved by measuring polymerase kinetics in Pacbio [80] and ionic current signals in Nanopore [81].
Fig. 2.
Structural variations. Schematic representation of the five major types of SVs.
3.3. Genome assembly using Third Generation Sequencing
The ultralong, noisy reads produced by TGS required the adaptation of assembly methods. At present, the two most popular softwares for de novo genome assembly of TGS data are FALCON and Canu. FALCON was developed directly by PacBio and published in 2013 [82]. It relies on a Hierarchical Genome Assembly Process (HGAP) where the longest reads are initially selected and act as ‘seed’ reads for the alignment of shorter reads. Reads are then ‘corrected’ to account for the high base calling error rate of TGS using the consensus sequence derived from the alignment. These ‘pre-assembled’ reads are then overlapped to each other using de Brujin graphs to generate the final contigs (Fig. 3 and Box 3). FALCON is diploid-aware and can generate alternative contigs representing the second haplotype out of the assembly graph. Three years later PacBio released FALCON-Unzip, an improved diploid-aware assembler that can take advantage of the heterozygous SNPs identified in the initial assembly to provide highly phased divergent haplotypes [83]. Today Canu is a very popular alternative. It was derived from the Celera Assembler in 2015 [84]. The latter was initially adapted to PacBio sequencing [85], demonstrating finished bacterial genomes [46] before being dismissed. Similarly to the Celera Assembler, Canu is an Overlap-Layout-Consensus (OLC) based assembler [86], specialized in the assembly of TGS long reads (Box 3). As FALCON, reads are error-corrected prior to the assembly. Despite the considerable improvement over short reads, the resulting assemblies were still a mixture of parental haplotypes and repeats were often collapsed in a single sequence. To overcome these issues associated with the presence of both parental haplotypes in the raw reads, in 2018 a new version of the software was made available, TrioCanu. This improved version can take advantage of the parental information to produce fully phased haplotypes [38], [87]. In this approach, the paternal and maternal genomes are sequenced with Illumina short-reads, and these reads are used to ‘bin’ the long-reads of the child based on divergent SNPs prior to the assembly, which therefore takes place independently for the two haplotypes. Interestingly, while reference genome projects have historically selected inbred individuals to minimize heterozygosity and simplify the assembly, in TrioCanu assembly process a high level of heterozygosity is welcomed, as it promotes effective read binning. One practical limitation shared by FALCON and Canu also with other prominent TGS/long-reads genomes assemblers, as ABruijn [88], Miniasm [89], HINGE [90] and Marvel [44], is that computational time and costs are starting to exceed sequencing expenses. The optimization of computational efforts has become an active area of research and new promising de novo genome assemblers are currently being developed, as Wtdbg2 [91], Flye [92], Peregrine [93], Shasta [94]. Generally, these new assemblers appear faster than FALCON and Canu, however the quality of the final genome may not necessarily be as accurate.
Fig. 3.
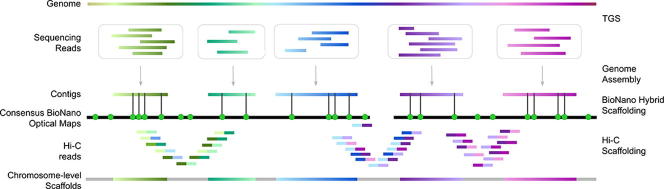
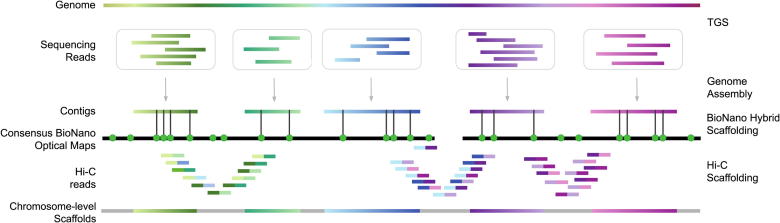
Chromosome-level scaffolding of de novo genome assemblies. Schematic illustration of an hybrid de novo genome assembly approach where linkage information obtained from optical and Hi-C maps is used to properly position contigs along the chromosomes. First, sequencing reads are assembled together to form contigs. Then, contigs are aligned to consensus BioNano optical maps, a process that enables to accurately order and orient the contigs with respect to each others and assign each contig to a specific chromosome. Here, BioNano optical maps are represented by black lines and the green dots indicate the labeled sequence motifs. Further ordering of the resulting scaffolds is made possible by Hi-C maps, that take into account the interactions between contiguous genomic loci. In the resulting chromosome-level scaffold colored segments represent genomic regions of known sequence while the remaining gaps between the scaffolds of unknown sequence are depicted as gray lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Box 3. Bioinformatics.
The continuous advancements experienced by sequencing technologies in the last 70 years, and the resulting exponential increase in the amount of sequencing data available, have been paralleled by improvements in data analysis tools. In the 1980s, the advent of information technology laid the foundations of bioinformatics. Processor computing allowed to automate the general principles of overlapping sequences by similarity using dedicated computer programs [16], [161], the first genome assemblers. In 1980, to describe the data obtained after assembly of shotgun sequencing reads, Staden coined the word ‘contig’. In his words, “a contig is a set of gel readings that are related to one another by overlap of their sequences. […] The gel readings in a contig can be summed to form a contiguous consensus sequence and the length of this sequence is the length of the contig” [161]. This is probably the first outline of the general principle of Overlap-Layout-Consensus (OLC) used to infer the original sequence from a subset of sequences. Since brute force alignment (i.e. generate all possible alignments and pick the one most likely to be true) was proven nearly impossible to solve even for short sequences, several overlap algorithms were implemented, including the Needleman–Wunsch algorithm for global alignments [162] and the ‘edit’ or Levenshtein distance [163]. Others were newly developed, such as those for local alignment [164]. Starting from 1985, the National Center for Biotechnology Information (NCBI) released its Basic Local Alignment Search Tool (BLAST) [165], which considerably speeded up the process of sequence alignment. The increased throughput of sequencers combined with the shotgun approach allowed to tackle more complex genomes. Venter’s team at TIGR developed a new software, named TIGR Assembler, that generated the whole genome of H. Influenzae out of approximately 24,000 DNA fragments using 30 CPU hours with half a gigabyte of RAM [166]. As the complexity of genomes was progressively understood, more refined tools were made available: RepeatMasker in 1996, to deal with repeated genomic regions [167], and GENSCAN in 1997, to predict gene structures [168]. In 1998, Green developed Phred, an algorithm that is still widely used today, along with two associated softwares for phred output analysis, phrap and consed [169]. Phred assigns a Quality Value (QV) score to each base called using the formula QV = −10 * log10 (Pe) where Pe is the probability of erroneous base-calling (e.g. QV20 implies 99% accuracy while QV30 implies 99.9% accuracy). Phred introduced reliable quality metrics for base calling, providing support to automated read analyses, particularly in repeated sequences [170].
The challenges associated with the human genome (Box 2) further fostered research in the field of genome assembly to an unprecedented scale. This is especially true for the effort led by Venter, that aimed at reconstructing the sequence of the human genome without BAC intermediates. After the release of the human genome assemblies, in 2002 two new online repositories were created in addition to Genbank to host human genome data, the University of California Santa Cruz (UCSC) Genome Browser [171] and Ensembl [172]. The unprecedented flood of sequencing data was barely accompanied by advancements in bioinformatic tools to store, process, analyze and visualize them. A new series of integrated open-source software and algorithms were released, including a radically new assembly approach based on de Bruijn graphs (DBG) [173]. Contrary to OLC, DBG assembly is counterintuitive, as it first chop the reads into shorter k-mers and then uses the k-mers to form a DBG from which the genome sequence is derived [174]. The first software using this approach was EULER in 2001 [175]. Other major milestones included short-read and vertebrate-specific aligner BLAT [176]; the platform for integrated genome analysis Galaxy [177]; the NCBI Short Read Archive (SRA) to store NGS raw data (2005); the assembly algorithms ALLPATHS [178], Velvet [179] and SOAPdenovo [180], all based on De Bruijn graphs; more efficient read alignment algorithms as Bowtie [181] and BWA [182]; integrated tools for read data management as SAMtools [183]; general algorithms for variant discovery as GATK [184] and Freebayes [185].
All genome assembly approaches and softwares have strengths and weaknesses, and practical considerations may be applied for choosing the appropriate approach, including the type and quality of input reads, coverage and genome size [95].
3.4. Scaffolding of genome assemblies
For large genomes, even long reads fail to generate end-to-end chromosome sequences, requiring linkage information to orient and order the contigs, a process known as scaffolding. Scaffolds comprise multiple contigs in their putative order on chromosomes, separated by genomic regions of undetermined sequence (represented by runs of Ns). One leading single-molecule DNA technology relevant for genome scaffolding is Bionano optical mapping, commercialized by Bionano Genomics (BioNano). The method is based on DNA labeling rather than sequencing and it aims at generating genetic optical maps. Optical maps were first described for yeast in the 1990s [96]. In the early approach, DNA molecules were stretched and fixed in agarose gel, resident restriction endonucleases were activated by Mg++ addition, and the differential fluorescence patterns between cleaved and intact sites were assessed using a microscope. In 2003, the technique was refined using restriction enzymes directed at specific recognition sites. T7 exonuclease was used to open single-strand gaps at those sites and gaps were filled with nucleotides labeled with fluorescent tags [97]. The current technology relies on the isolation of DNA fragments >150 Kbp and staining of the filaments using restriction enzymes recognizing specific 6–7 bp sequence motifs. DNA molecules are then stretched into nanoscale channels (NanoChannels) through progressively smaller sievers and imaged with a high-resolution camera [98]. Information from individual DNA molecules is built into consensus maps (cmaps) using the software Bionano Solve. The distances between motif-specific patterns represent the optical maps [98]. The linkage information provided by these maps is crucial to improve the process of de novo genome assembly [98], [99], potentially identifying and resolving misassemblies [100], [101], [102]. Draft assemblies generated using either NGS or TGS sequencing data alone are matched with consensus optical maps. The overlaps allow to orient and order the contigs, resolve chimeric joins, and estimate the length of gaps between adjacent contigs. This produces longer scaffolds, potentially spanning entire chromosomes [43], [103] (Fig. 3). Given the improved contiguity of hybrid genome assemblies generated using sequencing data in combination with optical mapping, this approach has been recently adopted by many de novo human [43], [104], [105], non-human [44], [106], [107], and plant [108] genome assembly projects (Fig. 1). In addition to its relevance for genome assembly, optical mapping allows the detection of SVs exceeding the length of individual NGS or long reads, by comparing the results to a reference genome [109]. Moreover, optical mapping was demonstrated to efficiently identify genome-wide methylation patterns, through methylation-sensitive restriction enzymes [110]. Until 2018, two enzymes were available for the staining of the DNA molecules: Nt.BspQI, with recognition sequence GCTCTTCN^ and Nb.BssSI with recognition sequence CACGA^G. These were both nicking enzymes that labeled DNA molecules introducing the fluorescent tag via a nick on one of the two DNA strands. This approach is collectively referred as Nick, Label, Repair and Stain (NLRS). In 2018, BioNano introduced the Direct Label and Stain (DLS) approach based on the enzyme DLE-1, with recognition sequence CTTAAG [106]. This enzyme can label the DNA without nicking it, considerably increasing the average length of the molecules that are stained and visualized. Currently, this is the only non-nicking enzyme available, however BioNano claims to release several more over the next few years.
Another popular scaffolding approach is Hi-C, a Chromosome Conformation Capture (3C) based technique that generates chromatin proximity information for all vs all regions of the genome [111]. This spatial information can be used to arrange contigs and scaffolds in a linear, potentially chromosome-level, sequence [112], [186]. In Hi-C sample preparation, cells are embedded in a matrix and the layers surrounding chromosomes are progressively washed away, thus preserving chromosome folding information. The DNA is then treated with a restriction enzyme and re-ligated to form new bonds with different, but spatially close, DNA molecules. The resulting library is sequenced with Illumina short reads and the obtained ‘map’ of DNA interactions is used to assign contigs and scaffolds to chromosomes [113], [114] (Fig. 3). While Hi-C currently appears to be the only approach enabling chromosome-level scaffolding in large genomes, it is usually less conservative than BioNano scaffolding. The unpredictable folding of chromatin resulting in long-range interactions of distant regions of chromosomes can lead to misassembly errors such as artificial inversions, scaffold misplacement within the same chromosome, or scaffold misassignment to different chromosomes [112]. These errors are better corrected combining different techniques and therefore several projects have started to adopt complementary scaffolding strategies [115], [116] (Fig. 1), even potentially allowing telomere-to-telomere assembly of entire chromosomes without gaps [48].
4. Summary and outlook
The way scientists generate genome assemblies has been and is constantly evolving. Through the constant refinement of existing technologies and the introduction of radically new DNA sequencing approaches and bioinformatic tools, the quality of the assemblies has never ceased to improve. The high-throughput capacity introduced by NGS and the enhanced quality offered by TGS have finally made also complex genomes accessible for whole-genome investigation. Human genetics research, including population genomics, genetic diseases mapping and diagnostics, personalized medicine programs, cancer research and prenatal testing have already benefited from the advancements in genome sequencing and assembly of the last decade. Similarly, these approaches are being increasingly employed in non-model organisms to understand ecological and evolutionary processes. The commitment to reference genome sequencing and assembly has now scaled up from single-species projects to multiple-species coordinated efforts, and projects aimed to produce high-quality genomes for most organisms using a combination of NGS and TGS approaches are currently underway.
Acknowledgments
Acknowledgments
We apologize to those whose work we were unable to discuss and cite in this review due to space constraints. We thank Erich Jarvis for critical reading of the manuscript and inputs. We also thank the two anonymous reviewers for providing insightful comments that helped improve and clarify this manuscript.
Declaration of interest
G.F. had one travel sponsored by Bionano Genomics. A.M.G., G.R.G. and L.G. declare no conflicts of interest.
Contributor Information
Alice Maria Giani, Email: amg4001@med.cornell.edu.
Guido Roberto Gallo, Email: guido.gallo@guest.unimi.it.
Luca Gianfranceschi, Email: luca.gianfranceschi@unimi.it.
Giulio Formenti, Email: gformenti@rockefeller.edu.
References
- 1.Watson J.D., Crick F.H.C. Molecular structure of nucleic acids. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F., Thompson E.O.P. The amino-acid sequence in the glycyl chain of insulin. II. The investigation of peptides from enzymic hydrolysates. Biochem J. 1953;53:366–374. doi: 10.1042/bj0530366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanger F., Thompson E.O.P. The amino-acid sequence in the glycyl chain of insulin. I. The identification of lower peptides from partial hydrolysates. Biochem J. 1953;53:353–366. doi: 10.1042/bj0530353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holley R.W., Apgar J., Everett G.A., Madison J.T., Marquisee M. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 5.Wu R., Kaiser A.D. Structure and base sequence in the cohesive ends of bacteriophage lambda DNA. J Mol Biol. 1968;35:523–537. doi: 10.1016/s0022-2836(68)80012-9. [DOI] [PubMed] [Google Scholar]
- 6.Brownlee G.G., Sanger F., Barrell B.G. The sequence of 5 s ribosomal ribonucleic acid. J Mol Biol. 1968;34:379–412. doi: 10.1016/0022-2836(68)90168-x. [DOI] [PubMed] [Google Scholar]
- 7.Min Jou W., Haegeman G., Ysebaert M., Fiers W. Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein. Nature. 1972;237:82–88. doi: 10.1038/237082a0. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert W., Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci USA. 1973;70:3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- 10.Sanger F., Coulson A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 11.Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975;14:3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- 12.Sanger F., Air G.M., Barrell B.G., Brown N.L., Coulson A.R. Nucleotide sequence of bacteriophage φX174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 13.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atkinson M.R., Deutscher M.P., Kornberg A., Russell A.F., Moffatt J.G. Enzymatic synthesis of deoxyribonucleic acid. XXXIV. Termination of chain growth by a 2′,3′-dideoxyribonucleotide. Biochemistry. 1969;8:4897–4904. doi: 10.1021/bi00840a037. [DOI] [PubMed] [Google Scholar]
- 15.Maxam A.M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staden R. A strategy of DNA sequencing employing computer programs. Nucl Acids Res. 1979;6:2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messing J., Crea R., Seeburg P.H. A system for shotgun DNA sequencing. Nucl Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanger F., Coulson A.R., Hong G.F., Hill D.F., Petersen G.B. Nucleotide sequence of bacteriophage λ DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 19.Baer R., Bankier A.T., Biggin M.D., Deininger P.L., Farrell P.J. DNA sequence and expression of the B95–8 Epstein—Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 20.GenBank and WGS Statistics n.d. https://www.ncbi.nlm.nih.gov/genbank/statistics/ (accessed August 11, 2019).
- 21.Sanger F. Sequences, sequences, and sequences. Annu Rev Biochem. 1988;57:1–28. doi: 10.1146/annurev.bi.57.070188.000245. [DOI] [PubMed] [Google Scholar]
- 22.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 23.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 24.Heather J.M., Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107:1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Head S.R., Komori H.K., LaMere S.A., Whisenant T., Van Nieuwerburgh F. Library construction for next-generation sequencing: overviews and challenges. Biotechniques. 2014;56 doi: 10.2144/000114133. 61–4, 66, 68, passim. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 29.Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorisson G.A., Smith A.V., Krishnan L., Stein L.D. The international hapmap project web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy S., Sutton G., Ng P.C., Feuk L., Halpern A.L. The diploid genome sequence of an individual human. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheeler D.A., Srinivasan M., Egholm M., Shen Y., Chen L. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 33.Worthey E.A., Mayer A.N., Syverson G.D., Helbling D., Bonacci B.B. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 34.1000 Genomes Project Consortium, Abecasis G.R., Altshuler D., Auton A., Brooks L.D. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reardon S. Giant study poses DNA data-sharing dilemma. Nature. 2015;525:16–17. doi: 10.1038/525016a. [DOI] [PubMed] [Google Scholar]
- 36.Alkan C., Sajjadian S., Eichler E.E. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaisson M.J.P., Huddleston J., Dennis M.Y., Sudmant P.H., Malig M. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korlach J., Gedman G., Kingan S.B., Chin C., Howard J.T. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. GigaScience. 2017;6:1–16. doi: 10.1093/gigascience/gix085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Souza R.d.C.M., Iraola G. D-VFPSGG. Nanopore sequencing significantly improves genome assembly of the protozoan parasite Trypanosoma cruzi. Genome Biol Evol. 2019;11:1952–1957. doi: 10.1093/gbe/evz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 41.de Koning A.P.J., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng G.X.Y., Lau B.T., Schnall-Levin M., Jarosz M., Bell J.M. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34:303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mostovoy Y., Levy-Sakin M., Lam J., Lam E.T., Hastie A.R. A hybrid approach for de novo human genome sequence assembly and phasing. Nat Methods. 2016;13:587–590. doi: 10.1038/nmeth.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowoshilow S., Schloissnig S., Fei J.-F., Dahl A., Pang A.W.C. The axolotl genome and the evolution of key tissue formation regulators. Nature. 2018;554:50–55. doi: 10.1038/nature25458. [DOI] [PubMed] [Google Scholar]
- 45.Biscotti M.A., Olmo E., Heslop-Harrison J.S.P. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015;23:415–420. doi: 10.1007/s10577-015-9499-z. [DOI] [PubMed] [Google Scholar]
- 46.Koren S., Harhay G.P., Smith T.P.L., Bono J.L., Harhay D.M. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14:R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon D., Huddleston J., Chaisson M.J.P., Hill C.M. Long-read sequence assembly of the gorilla genome. Science. 2016;352:aae0344. doi: 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A. Telomere-to-telomere assembly of a complete human X chromosome. bioRxiv. 2019:735928. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braslavsky I., Hebert B., Kartalov E., Quake S.R. Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris T.D., Buzby P.R., Babcock H., Beer E., Bowers J. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 51.Deamer D., Akeson M., Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34:518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Travers K.J., Chin C.-S., Rank D.R., Eid J.S., Turner S.W. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucl Acids Res. 2010;38 doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levene M.J., Korlach J., Turner S.W., Foquet M., Craighead H.G. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 54.Eid J., Fehr A., Gray J., Luong K., Lyle J. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 55.Carneiro M.O., Russ C., Ross M.G., Gabriel S.B., Nusbaum C. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics. 2012;13:375. doi: 10.1186/1471-2164-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L., Li Y., Li S., Hu N., He Y. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizuguchi T., Toyota T., Adachi H., Miyak N., Matsumoto N. Detecting a long insertion variant in SAMD12 by SMRT sequencing: implications of long-read whole-genome sequencing for repeat expansion diseases. J Hum Genet. 2019;64:191–197. doi: 10.1038/s10038-018-0551-7. [DOI] [PubMed] [Google Scholar]
- 58.Rhoads A., Au K.F. PacBio sequencing and its applications. Genomics Proteomics Bioinf. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wenger A.M., Peluso P., Rowell W.J., Chang P.C., Hall R.J. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang S., He J., Chang S., Zhang P., Liang F. Identifying single bases in a DNA oligomer with electron tunnelling. Nat Nanotechnol. 2010;5:868–873. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cherf G.M., Lieberman K.R., Rashid H., Lam C.E., Karplus K. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manrao E.A., Derrington I.M., Laszlo A.H., Langford K.W., Hopper M.K. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Payne A., Holmes N., Rakyan V., Loose M. BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics. 2019;35:2193–2198. doi: 10.1093/bioinformatics/bty841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Istace B., Friedrich A., d’Agata L., Faye S., Payen E. de novo assembly and population genomic survey of natural yeast isolates with the Oxford Nanopore MinION sequencer. GigaScience. 2017;6:1–13. doi: 10.1093/gigascience/giw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.New “R10” nanopore released into early access. Oxford nanopore technologies https://nanoporetech.com/about-us/news/new-r10-nanopore-released-early-access (accessed August 18, 2019).
- 67.Giordano F., Aigrain L., Quail M.A., Coupland P., Bonfield J.K. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms. Sci Rep. 2017;7:3935. doi: 10.1038/s41598-017-03996-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roberts R.J., Carneiro M.O., Schatz M.C. The advantages of SMRT sequencing. Genome Biol. 2013;14:405. doi: 10.1186/gb-2013-14-7-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuleshov V., Xie D., Chen R., Pushkarev D., Ma Z. Whole-genome haplotyping using long reads and statistical methods. Nat Biotechnol. 2014;32:261–266. doi: 10.1038/nbt.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Merker J.D., Wenger A.M., Sneddon T., Grove M., Zappala Z. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet Med. 2018;20:159–163. doi: 10.1038/gim.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alkan C., Coe B.P., Eichler E.E. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 75.Eichler E.E., Flint J., Gibson G., Kong A., Leal S.M. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J., Wurm Y., Nipitwattanaphon M., Riba-Grognuz O., Huang Y.-C. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;493:664–668. doi: 10.1038/nature11832. [DOI] [PubMed] [Google Scholar]
- 77.Nishikawa H., Iijima T., Kajitani R., Yamaguchi J., Ando T. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet. 2015;47:405–409. doi: 10.1038/ng.3241. [DOI] [PubMed] [Google Scholar]
- 78.Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 80.Schadt E.E., Banerjee O., Fang G., Feng Z., Wong W.H. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 2013;23:129–141. doi: 10.1101/gr.136739.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rand A.C., Jain M., Eizenga J.M., Musselman-Brown A., Olsen H.E. Mapping DNA methylation with high-throughput nanopore sequencing. Nat Methods. 2017;14:411–413. doi: 10.1038/nmeth.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chin C., Alexander D.H., Marks P., Klammer A.A., Drake J. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 83.Chin C., Peluso P., Sedlazeck F.J., Nattestad M., Concepcion G.T. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berlin K., Koren S., Chin C., Drake J., Landolin J.M. Assembling large genomes with single-molecule sequencing and locality sensitive hashing. Nat Biotechnol. 2015;33:623–630. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 85.Koren S., Schatz M.C., Walenz B.P., Martin J., Howard J.T. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koren S., Rhie A., Walenz B.P., Dilthey A.T., Bickhart D.M. De novo assembly of haplotype-resolved genomes with trio binning. Nat Biotechnol. 2018;36:1174–1182. doi: 10.1038/nbt.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin Y., Yuan J., Kolmogorov M., Shen M.W., Chaisson M. Assembly of long error-prone reads using de Bruijn graphs. Proc Natl Acad Sci USA. 2016;113:E8396–E8405. doi: 10.1073/pnas.1604560113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016;32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kamath G.M., Shomorony I., Xia F., Courtade T.A., Tse D.N. HINGE: long-read assembly achieves optimal repeat resolution. Genome Res. 2017;27:747–756. doi: 10.1101/gr.216465.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruan J., Li H. Fast and accurate long-read assembly with wtdbg2. bioRxiv. 2019:530972. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 93.Chin C.-S., Khalak A. Human Genome Assembly in 100 Minutes. bioRxiv. 2019:705616. [Google Scholar]
- 94.Shafin K, Pesout T, Lorig-Roach R, Haukness M, Olsen HE, et al. Efficient de novo assembly of eleven human genomes using PromethION sequencing and a novel nanopore toolkit. bioRxiv 2019:561. [DOI] [PMC free article] [PubMed]
- 95.Dominguez Del Angel V, Hjerde E, Sterck L, Capella-Gutierrez S, Notredame C, et al. Ten steps to get started in genome assembly and annotation. F1000Res 2018; 7. pii: ELIXIR-148. [DOI] [PMC free article] [PubMed]
- 96.Schwartz D.C., Li X., Hernandez L.I., Ramnarain S.P., Huff E.J. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 97.Ramanathan A., Huff E.J., Lamers C.C., Potamousis K.D., Forrest D.K. An integrative approach for the optical sequencing of single DNA molecules. Anal Biochem. 2004;330:227–241. doi: 10.1016/j.ab.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 98.Lam E.T., Hastie A., Lin C., Ehrlich D., Das S.K. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Teague B., Waterman M.S., Goldstein S., Potamousis K., Zhou S. High-resolution human genome structure by single-molecule analysis. Proc Natl Acad Sci USA. 2010;107:10848–10853. doi: 10.1073/pnas.0914638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nagarajan N., Read T.D., Pop M. Scaffolding and validation of bacterial genome assemblies using optical restriction maps. Bioinformatics. 2008;24:1229–1235. doi: 10.1093/bioinformatics/btn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Howe K., Wood J.M.D. Using optical mapping data for the improvement of vertebrate genome assemblies. GigaScience. 2015;4:10. doi: 10.1186/s13742-015-0052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tang H., Lyons E., Town C.D. Optical mapping in plant comparative genomics. GigaScience. 2015;4:3. doi: 10.1186/s13742-015-0044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hastie A.R., Dong L., Smith A., Finklestein J., Lam E.T. Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex Aegilops tauschii genome. PLoS One. 2013;8 doi: 10.1371/journal.pone.0055864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pendleton M., Sebra R., Pang A.W.C., Ummat A., Franzen O. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods. 2015;12:780–786. doi: 10.1038/nmeth.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seo J.-S., Rhie A., Kim J., Lee S., Sohn M.-H. De novo assembly and phasing of a Korean human genome. Nature. 2016;538:243–247. doi: 10.1038/nature20098. [DOI] [PubMed] [Google Scholar]
- 106.Formenti G., Chiara M., Poveda L., Francoijs K.-J., Bonisoli-Alquati A. SMRT long reads and direct label and Stain optical maps allow the generation of a high-quality genome assembly for the European barn swallow (Hirundo rustica rustica) GigaScience. 2019;8(1) doi: 10.1093/gigascience/giy142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lind A.L., Lai Y.Y.Y., Mostovoy Y., Holloway A.K., Iannucci A. Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards. Nat Ecol Evol. 2019;3:1241–1252. doi: 10.1038/s41559-019-0945-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiao Y., Peluso P., Shi J., Liang T., Stitzer M.C. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524–527. doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mak A.C.Y., Lai Y.Y.Y., Lam E.T., Kwok T.-P., Leung A.K.Y. Genome-wide structural variation detection by genome mapping on nanochannel arrays. Genetics. 2016;202:351–362. doi: 10.1534/genetics.115.183483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ananiev G.E., Goldstein S., Runnheim R., Forrest D.K., Zhou S. Optical mapping discerns genome wide DNA methylation profiles. BMC Mol Biol. 2008;9:68. doi: 10.1186/1471-2199-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dudchenko O., Batra S.S., Omer A.D., Nyquist S.K., Hoeger M. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Teh B.T., Lim K., Yong C.H., Ng C.C.Y., Rao S.R. The draft genome of tropical fruit durian (Durio zibethinus) Nat Genet. 2017;49:1633–1641. doi: 10.1038/ng.3972. [DOI] [PubMed] [Google Scholar]
- 115.Bickhart D.M., Rosen B.D., Koren S., Sayre B.L., Hastie A.R. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 2017;49:643–650. doi: 10.1038/ng.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wallberg A., Bunikis I., Pettersson O.V., Mosbech M.-B., Childers A.K. A hybrid de novo genome assembly of the honeybee, Apis mellifera, with chromosome-length scaffolds. BMC Genomics. 2019;20:275. doi: 10.1186/s12864-019-5642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith L.M., Fung S., Hunkapiller M.W., Hunkapiller T.J., Hood L.E. The synthesis of oligonucleotides containing an aliphatic amino group at the 5′ terminus: synthesis of fluorescent DNA primers for use in DNA sequence analysis. Nucl Acids Res. 1985;13:2399–2412. doi: 10.1093/nar/13.7.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smith L.M., Sanders J.Z., Kaiser R.J., Hughes P., Dodd C. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 119.Prober J.M., Trainor G.L., Dam R.J., Hobbs F.W., Robertson C.W. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987;238:336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- 120.Tabor S., Richardson C.C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee L.G., Connell C.R., Woo S.L., Cheng R.D., McArdle B.F. DNA sequencing with dye-labeled terminators and T7 DNA polymerase: effect of dyes and dNTPs on incorporation of dye-terminators and probability analysis of termination fragments. Nucl Acids Res. 1992;20:2471–2483. doi: 10.1093/nar/20.10.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Murray V. Improved double-stranded DNA sequencing using the linear polymerase chain reaction. Nucl Acids Res. 1989;17:8889. doi: 10.1093/nar/17.21.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Metzker M.L., Lu J., Gibbs R.A. Electrophoretically uniform fluorescent dyes for automated DNA sequencing. Science. 1996;271:1420–1422. doi: 10.1126/science.271.5254.1420. [DOI] [PubMed] [Google Scholar]
- 124.DeAngelis M.M., Wang D.G., Hawkins T.L. Solid-phase reversible immobilization for the isolation of PCR products. Nucl Acids Res. 1995;23:4742–4743. doi: 10.1093/nar/23.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang J., Fang Y., Hou J.Y., Ren H.J., Jiang R. Use of non-cross-linked polyacrylamide for four-color DNA sequencing by capillary electrophoresis separation of fragments up to 640 bases in length in two hours. Anal Chem. 1995;67:4589–4593. doi: 10.1021/ac00120a026. [DOI] [PubMed] [Google Scholar]
- 126.Salas-Solano O., Carrilho E., Kotler L., Miller A.W., Goetzinger W. Routine DNA sequencing of 1000 bases in less than one hour by capillary electrophoresis with replaceable linear polyacrylamide solutions. Anal Chem. 1998;70:3996–4003. doi: 10.1021/ac980457f. [DOI] [PubMed] [Google Scholar]
- 127.Hyman E.D. A new method of sequencing DNA. Anal Biochem. 1988;174:423–436. doi: 10.1016/0003-2697(88)90041-3. [DOI] [PubMed] [Google Scholar]
- 128.Ronaghi M., Karamohamed S., Pettersson B., Uhlén M., Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 129.Nyrén P. Enzymatic method for continuous monitoring of DNA polymerase activity. Anal Biochem. 1987;167:235–238. doi: 10.1016/0003-2697(87)90158-8. [DOI] [PubMed] [Google Scholar]
- 130.Ronaghi M., Uhlén M., Nyrén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 131.Tawfik D.S., Griffiths A.D. Man-made cell-like compartments for molecular evolution. Nat Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- 132.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mitra R.D., Shendure J., Olejnik J., Edyta-Krzymanska-Olejnik, Church G.M. Fluorescent in situ sequencing on polymerase colonies. Anal Biochem. 2003;320:55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 134.Kawashima E, Farinelli L, Mayer P. 1998, Method of nucleic acid amplification, WO1998044151A1.
- 135.Mitra R.D., Church G.M. In situ localized amplification and contact replication of many individual DNA molecules. Nucl Acids Res. 1999;27 doi: 10.1093/nar/27.24.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Adessi C., Matton G., Ayala G., Turcatti G., Mermod J.J. Solid phase DNA amplification: characterisation of primer attachment and amplification mechanisms. Nucl Acids Res. 2000;28:E87. doi: 10.1093/nar/28.20.e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ost TB. 2006, Improved Polymerases WO. 2006:120433. [Google Scholar]
- 138.Ruparel H., Bi L., Li Z., Bai X., Kim D.H. Design and synthesis of a 3′-O-allyl photocleavable fluorescent nucleotide as a reversible terminator for DNA sequencing by synthesis. Proc Natl Acad Sci USA. 2005;102:5932–5937. doi: 10.1073/pnas.0501962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Seo T.S., Bai X., Kim D.H., Meng Q., Shi S. Four-color DNA sequencing by synthesis on a chip using photocleavable fluorescent nucleotides. Proc Natl Acad Sci USA. 2005;102:5926–5931. doi: 10.1073/pnas.0501965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D.H. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 142.Huang Y.-F., Chen S.-C., Chiang Y.-S., Chen T.-H., Chiu K.-P. Palindromic sequence impedes sequencing-by-ligation mechanism. BMC Syst Biol. 2012;6(Suppl. 2):S10. doi: 10.1186/1752-0509-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rothberg J.M., Hinz W., Rearick T.M., Schultz J., Mileski W. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 144.Loman N.J., Misra R.V., Dallman T.J., Constantinidou C., Gharbia S.E. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 145.Greenleaf W.J., Sidow A. The future of sequencing: convergence of intelligent design and market Darwinism. Genome Biol. 2014;15:303. doi: 10.1186/gb4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zimmerman E. Illumina. MIT Technology Review; 2014.
- 147.Hayden EC. Technology: The $1,000 genome. Nature. 2014;507:294–295. doi: 10.1038/507294a. [DOI] [PubMed] [Google Scholar]
- 148.Human genome project timeline of events. Genome.gov n.d. https://www.genome.gov/human-genome-project/Timeline-of-Events [accessed August 11, 2019].
- 149.Roberts L. Why Watson quit as project head. Science. 1992;256:301–302. doi: 10.1126/science.256.5055.301. [DOI] [PubMed] [Google Scholar]
- 150.Collins F., Galas D. A new five-year plan for the U.S. Human Genome Project. Science. 1993;262:43–46. doi: 10.1126/science.8211127. [DOI] [PubMed] [Google Scholar]
- 151.Murray J.C., Buetow K.H., Weber J.L., Ludwigsen S., Scherpbier-Heddema T. A comprehensive human linkage map with centimorgan density. Cooperative Human Linkage Center (CHLC) Science. 1994;265:2049–2054. doi: 10.1126/science.8091227. [DOI] [PubMed] [Google Scholar]
- 152.U.S. HGP on fast track for early completion n.d. https://web.ornl.gov/sci/techresources/Human_Genome/publicat/hgn/v10n1/hgn101_2.pdf [accessed August 20, 2019].
- 153.Collins F, Patrinos A, Jordan E, Chakravarti A, Gesteland R. New goals for the U.S. Human Genome Project: 1998-2003. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- 154.Weber J.L., Myers E.W. Human whole-genome shotgun sequencing. Genome Res. 1997;7:401–409. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]
- 155.Myers E.W., Sutton G.G., Delcher A.L., Dew I.M., Fasulo D.P. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 156.Two thirds of human DNA script deciphered by human genome project. Genome.gov n.d. https://www.genome.gov/10002080/2000-release-twothirds-human-dna-sequenced [accessed August 11, 2019].
- 157.Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J. DNA sequencing at 40: past, present and future. Nature. 2017;550:345–353. doi: 10.1038/nature24286. [DOI] [PubMed] [Google Scholar]
- 158.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 159.Schneider V.A., Graves-Lindsay T., Howe K., Bouk N., Chen H.-C. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–864. doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Human genome project FAQ. Genome.gov n.d. https://www.genome.gov/human-genome-project/Completion-FAQ [accessed August 11, 2019].
- 161.Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucl Acids Res. 1980;8:3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 163.Levenshtein V.I. Binary codes capable of correcting deletions, insertions, and reversals. Soviet physics doklady. 1966;10:707–710. [Google Scholar]
- 164.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 165.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 166.Sutton G.G., White O., Adams M.D., Kerlavage A.R. TIGR assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Technol. 1995;1:9–19. [Google Scholar]
- 167.Smit A.F. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucl Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 169.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 170.Ewing B, Hillier LD, Wendl MC, Green P. Base-calling of automated sequencer traces usingPhred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 171.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hubbard T., Barker D., Birney E., Cameron G., Chen Y. The Ensembl genome database project. Nucl Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Idury R.M.W.M.S. A new algorithm for DNA sequence assembly. J Comput Biol. 1995;2:291–306. doi: 10.1089/cmb.1995.2.291. [DOI] [PubMed] [Google Scholar]
- 174.Li Z., Chen Y., Mu D., Yuan J., Shi Y. Comparison of the two major classes of assembly algorithms: overlap-layout-consensus and de-bruijn-graph. Brief Funct Genomics. 2012;11:25–37. doi: 10.1093/bfgp/elr035. [DOI] [PubMed] [Google Scholar]
- 175.Pevzner P.A., Tang H., Waterman M.S. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kent W.J. BLAT—the BLAST-Like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Butler J., MacCallum I., Kleber M., Shlyakhter I.A., Belmonte M.K. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li R., Zhu H., Ruan J., Qian W., Fang X. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012;arXiv:1207.3907 [Google Scholar]
- 186.Kaplan N, Dekker J. High-throughput genome scaffolding from in vivo DNA interaction frequency. Nat. Biotechnol. 2013;31:1143–1147. doi: 10.1038/nbt.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]