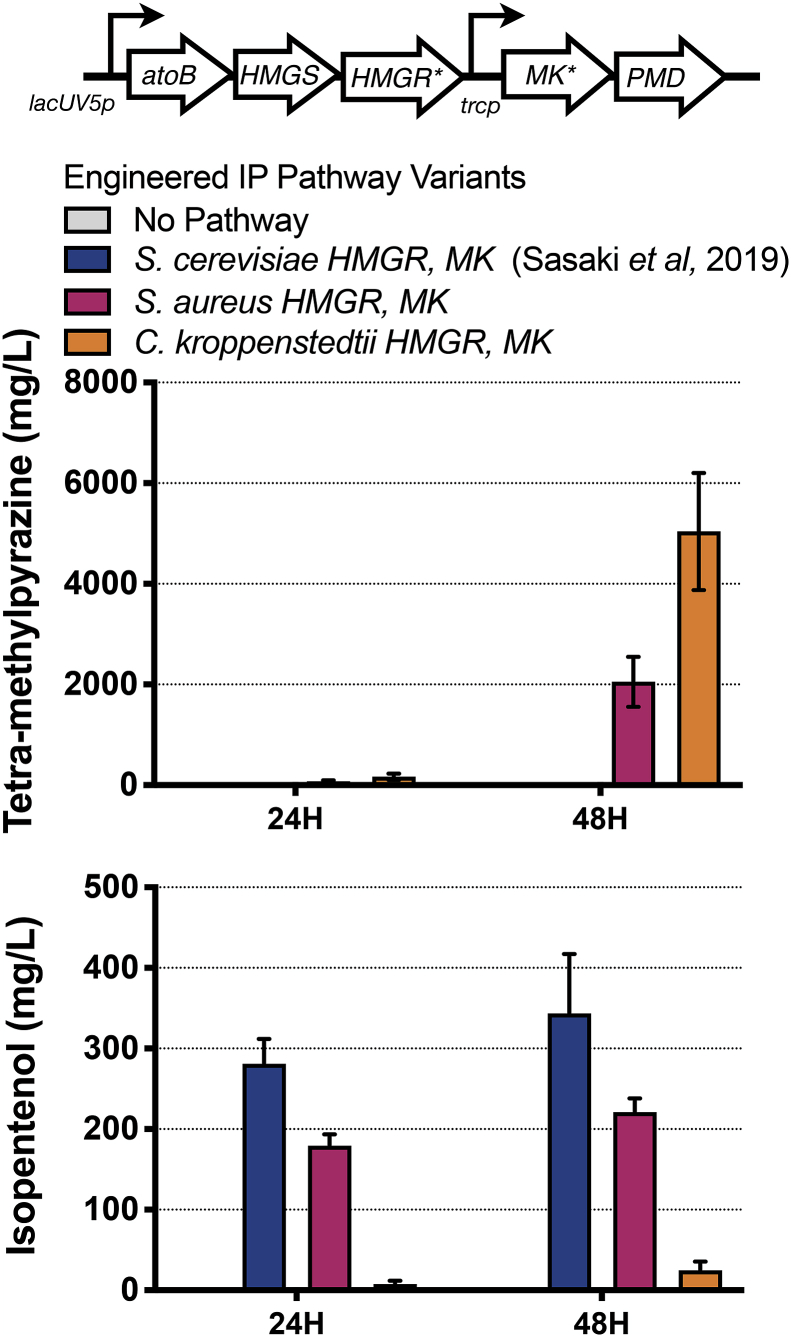
Fig. 2.
mk and hmgR Variants from bacterial species bias production towards tetra-methylpyrazine.
Top panel. Schematic of the heterologous isopentenol production pathway. The two genes selected for optimization by substitution with homologs are indicated with an asterisk (*). The proposed metabolic pathway for the conversion of glucose to isopentenol has been previously described in Sasaki et al (2019).
Bottom panels. Analysis of the engineered isopentenol production pathway in C. glutamicum using mk and hmgR homologs from S. aureus and C. kroppenstedtii in a 24 well plate format. C. glutamicum ΔpoxB ΔldhA strain harboring the original isopentenol production pathway (JBEI-19559) was compared with variants where the S. cerevisiae MK and HMGR were replaced with mk and hmgR from S. aureus (JBEI-19652) or C. kroppenstedtii (JBEI-19658). Samples were cultivated in CGXII media 4% d-glucose in a 24 well plate format. TMP or isopentenol titers were analyzed at the timepoints indicated and are an average of three biological replicates. The error bars represent standard error.