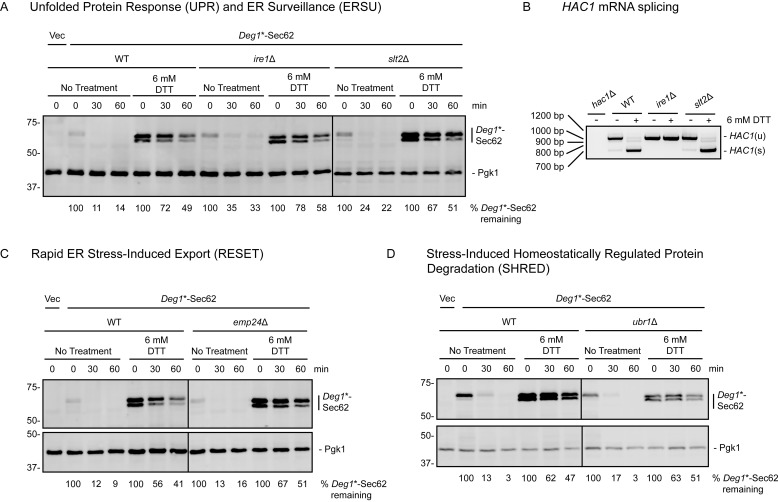
Figure 8.
Characterized ER stress-sensing pathways do not regulate ERAD-T in the presence or absence of ER stress. A, C, and D, cycloheximide chase analysis of yeast of the indicated genotypes expressing Deg1*-Sec62 or harboring an empty vector (Vec) cultured in the presence or absence of 6 mm DTT for 1 h. DTT was maintained at the same concentration during incubation with cycloheximide. Deg1*-Sec62 was detected with peroxidase anti-peroxidase antibodies. Pgk1 served as a loading control. The percentage of Deg1*-Sec62 remaining at each time point (normalized to Pgk1) is indicated below the images. IRE1 (A), SLT2 (A), EMP24 (C), and UBR1 (D) gene deletions were verified by PCR genotyping.8 B, yeast of the indicated genotypes were incubated in the presence or absence of 6 mm DTT for 1 h prior to RNA extraction and RT-PCR to analyze HAC1 mRNA splicing. Expected product sizes are 969 bp for unspliced HAC1 (HAC1(u)) and 717 bp for spliced HAC1 (HAC1(s)). Experiments depicted in A, C, and D were performed three times (with the exception of the right panel of A, which was performed two times). The experiment depicted in B was performed one time.