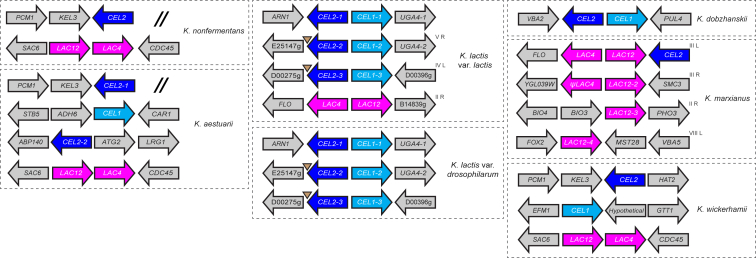
Figure 3.
Organization of LAC and CEL Genes in the Genomes of Kluyveromyces Species and Varieties
Dashed lines delimit the genes that belong to each species. The LAC genes are shown in magenta. CEL1 and CEL2 are shown in cyan and dark blue, respectively. Missing information due to incomplete genome assembly is indicated with a double slash symbol in the K. nonfermentans and K. aestuarii panels. LTR elements are represented by inverted triangles. The organization of the K. marxianus genes is based on the CBS397 assembly [7]. ψLAC4 is a conserved pseudogene and LAC12-4 is a pseudogene only in some strains, and variability both in the sequence of Lac4 and in the genomic organization in this telomere of chromosome 8 is seen between strains [9]. Superscript text indicates the position (chromosome number and arm) of the LAC and CEL genes in K. marxianus and the K. lactis var. lactis variants. Loci in K. lactis var lactis and K. lactis var. drosophilarum are syntenic, but reciprocal translocations may have changed chromosome numbers.
See also Figures S2 and S3 and Table S1.