Figure 1.
A Deficiency Screen Identified Two Overlapping Deletion Lines that Significantly Increased the Proportion of mt:yak
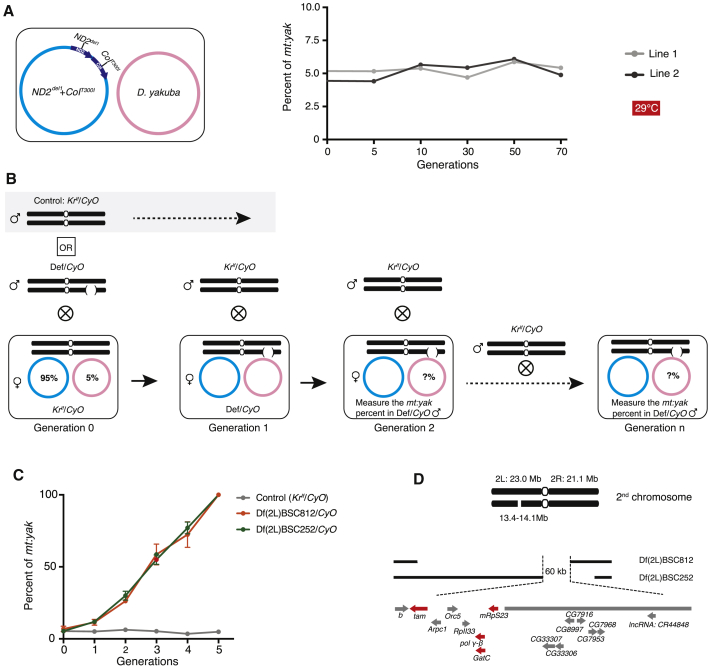
(A) The stable heteroplasmic line. The D. melanogaster mtDNA (blue circle) has two mutations, ND2del1 and CoIT300I. ND2del1 is a 9-bp in-frame deletion in the gene encoding NADH-dehydrogenase 2 (dark blue); it is a viable hypomorphic allele. CoIT300I is a temperature-sensitive allele of cytochrome c oxidase I (dark blue); when homoplasmic, it is lethal at 29°C but viable at lower temperatures. Although mt:yak (pink circle) is fully functional in D. melanogaster, it is usually outcompeted by the endogenous wild-type D. melanogaster mtDNA. However, when paired with mt:ND2del1+CoIT300I,mt:yak is stably transmitted at ∼5% for over 70 generations at 29°C [8].
(B) The genetic cross scheme to introduce deletion chromosomes (Table S1) into the stable heteroplasmic line (only shown for the 2nd chromosome deficiencies schematized by a bracketed interruption). First, 10 KrIf/CyO heteroplasmic females (generation 0) were crossed to 5 deficiency/CyO (Def/CyO) males to generate 10–20 female progeny with the genotype of Def/CyO (generation 1), which were further crossed to 10 KrIf/CyO males to produce progeny (generation 2). Def/CyO males of generation 2 were collected to measure the mt:yak percentage via qPCR. For some deficiencies, Def/CyO males were collected at multiple generations to follow the mt:yak percentage over time. In controls, KrIf/CyO males were used for the first cross instead of Def/CyO males. All the subsequent steps were the same, and KrIf/CyO males of generation 2 were collected to measure the mt:yak percentage.
(C) Two overlapping deficiencies (balanced by CyO) increased mt:yak transgenerationally (see also Tables S2 and S3). The mt:yak percentage reached 100% after five generations. Error bars indicate SDs of three independent experiments.
(D) The region deleted in both deficiencies contains 15 genes, four of which encode mitochondrial proteins (red) (also refer to Figure S1).