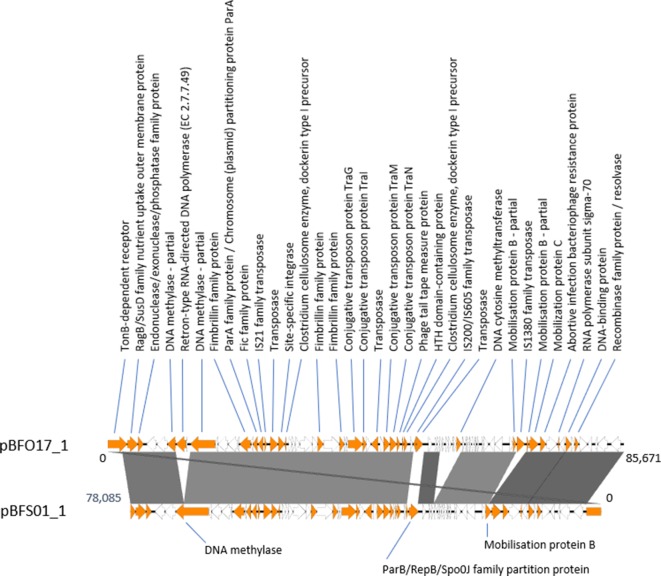
Fig. 3.
Linear representation of an alignment of putative circular plasmid sequences pBFO17_1 and pBFS01_1 (reverse complement for better visualization) using EasyFig [82]. EasyFig uses blast to identify sequences of similarity. Sequence similarities of >98 % are indicated by full colouring, a darker colour indicates a higher % ID. Products of annotated CDSs are shown. CDSs annotated as hypothetical or domain of unknown function are coloured white. The two sequences show a very high degree of similarity. pBFO17_1 is 7586 bp longer than pBFS01_1. This is mainly due to the insertion of a reverse transcriptase (pBFO17_1, 11367…13034) (disrupting a DNA methylase), the insertion of prophage (from position 56125 to 61162) (identified as an incomplete prophage using phaster [83] and an IS1380 family-like transposase (67933…69237). The regions pBFO17_1 50711…52501 and pBFS01_1 32248…30304 are not similar. Possibly, the insertion of two transposases in pBFO17_1 have excised most of the ParB-family DNA partitioning protein in the corresponding sequence range in pBFS01_1.