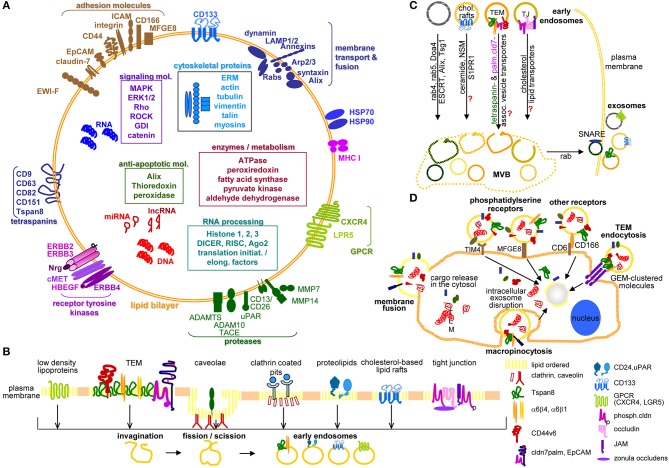
Figure 1.
Exosome characterization, biogenesis, and targeting. (A) Exosomes are composed of a lipid bilayer, transmembrane protein and the cytoplasm containing proteins, mRNA, non-coding RNA like miRNA and lncRNA and DNA, where PaCIC-TEX, express the CIC markers Tspan8, integrin α6β1/α6β4, CD44v6, CD133, CXCR4, LRP5, EpCAM, and cldn7. Other transmembrane proteins are linked to Exo biogenesis. (B) Exo biogenesis starts with the invagination of membrane microdomains that are characterized by ordered lipids, like low-density lipoprotein, caveolae, clathrin-coated pits, cholesterol-based lipid rafts, and others. (C) After fission and scission of invaginated membrane domains, the EE are guided toward MVB, the traffic differs between the origins from distinct lipid-enriched domains. Most abundant is rab4, rab5, Doa4 promoted migration and invagination into MVB via the ESCRT system. Components of cholesterol-based lipid raft-, TJ-, or TEM-derived EE are not completely explored. Guidance from MVB to the plasma membrane involves rab proteins, phospholipase D, and SNARE. (D) The contact between Exo and target cells can proceed via fusion of the Exo membrane with the cell membrane, by macropinocytosis, receptor ligand binding such as phosphatidylserine binding to TIM4 or MFGE8 or CD166 binding to CD6 or may be facilitated by Exo membrane protein complexes binding to invagination-prone complexes as described for TEM binding to the TCR complex. Exo also bind to the ECM or matricellular proteins, CD44 and integrins being most frequently involved. Full name of proteins are listed in Table S1. In brief, cells use a variety of pathways for the generation of EE, the traffic toward MVB, the loading of ILV with proteins, coding and non-coding RNA and DNA. Exo may preferentially bind to and be taken up by receptor-ligand binding, uptake being facilitated by the engagement of protein complexes at both the Exo and the target cell.