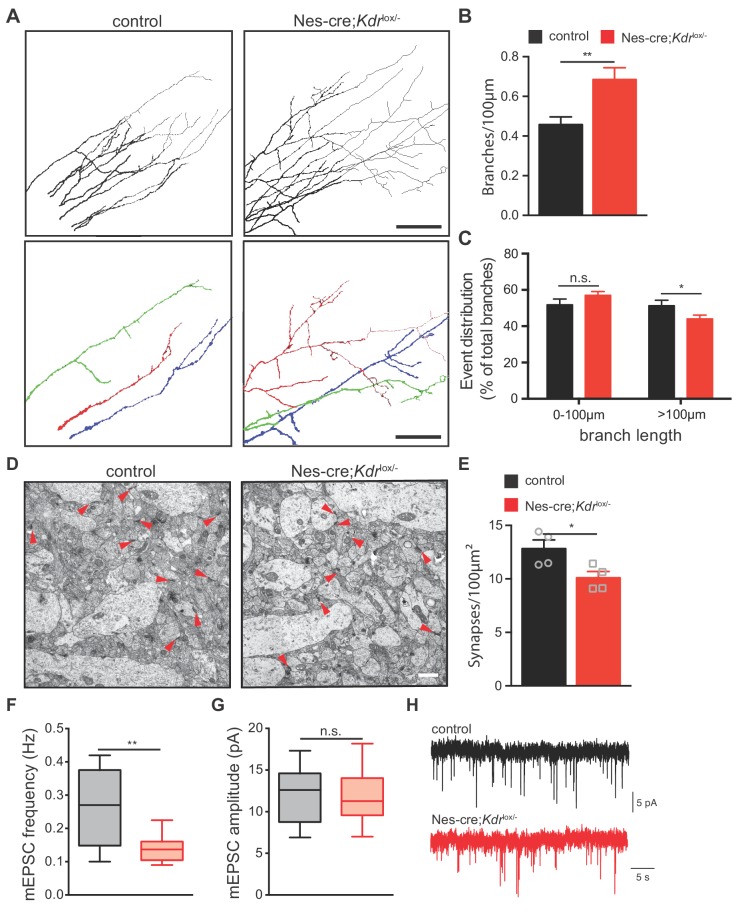
Figure 5. Nes-cre;Kdrlox/-mice show defects in axon branching and synapse density in vivo.
(A) Representative tracings in the CA1 stratum radiatum of CA3 DiI labeled axons from P10 control and Nes-cre;Kdrlox/- mice. Selected individual axon traces are shown highlighted in different colors in the panels below. Scale bar 100 µm. (B,C) Quantification of the axon branch number per 100 µm axon length (B) as well as their length distribution (C). Data are represented as mean ± SEM from n = 15 control and n = 18 Nes-cre;Kdrlox/- pups of 6 independent litters (B,C). n.s. not significant; *p<0.05; **p<0.01; unpaired Student’s ttest. (D) Representative electron micrographs of the hippocampal CA1 stratum radiatum from P10 control and Nes-cre;Kdrlox/- mice. Red arrowheads indicate synapses. Scale bar 1 µm. (E) Quantification of synapse density in control and Nes-cre;Kdrlox/- mice. Synapse numbers from 8 fields of view of n = 4 mice from each genotype were analyzed. *p<0.05; unpaired Student’s ttest. (F,G) Average mEPSC frequency (F) and amplitude (G) in control and Nes-cre;Kdrlox/- mice. n.s. not significant; **p<0.01; unpaired Student’s ttest. (H) Representative mEPSC recordings in control and Nes-cre;Kdrlox/- mice.