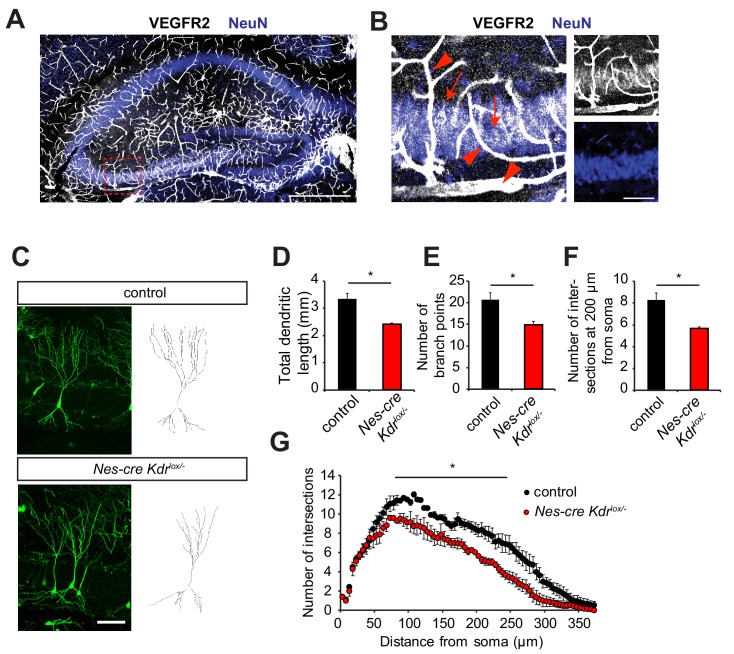
Figure 1. Nervous system specific VEGFR2 loss affects dendritic development in CA3 pyramidal neurons.
(A, B) Immunostainings for VEGFR2 revealed high levels of the receptor in the vessels (arrowheads) and double-labeling with the neuronal marker NeuN shows that the receptor is also expressed in pyramidal neurons (arrows) of the CA3 region in P10 hippocampus. Scale bar: A: 500 µm, B: 100 µm. (C–G) Nervous system specific deletion of VEGFR2 results in reduced dendritic arborization of CA3 pyramidal neurons. Nes-cre Kdrlox/- mice were crossed to Thy1-GFP transgenic mice to visualize whole morphology of pyramidal neurons. Z-projections of confocal images and the corresponding outlined tracings from CA3 neurons of P10 Nes-cre Kdrlox/- and control littermates are shown in (C). Total dendritic length (D), number of branch points (E) and the number of dendrites at 200 µm from the soma (F) were significantly reduced in Nes-cre Kdrlox/- mice compared to control littermates. 3D Sholl analysis of confocal z-stacks shows reduced branching and dendritic complexity of Nes-cre Kdrlox/- CA3 pyramidal neurons (G). Scale bar: 100 µm. n = 3 mice per genotype; SEM; *p<0.05.