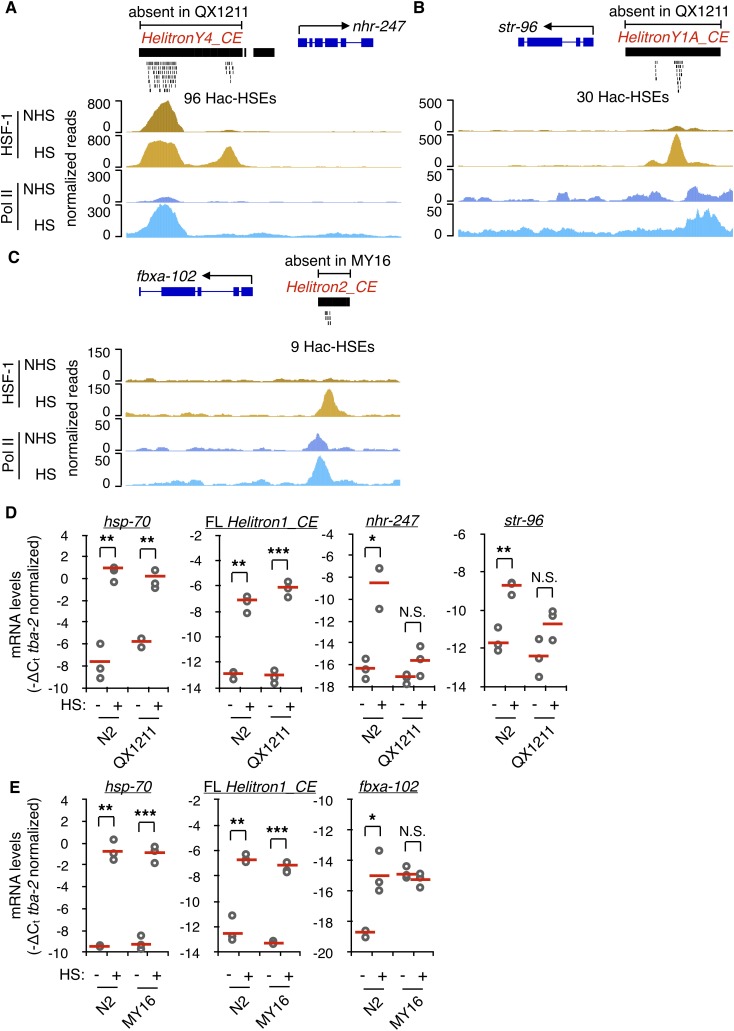
Figure 5. Hac-HSEs diversify the HSR in C. elegans.
(A) Genome browser screenshot depicting the region surrounding the Hac-HSE HSR gene nhr-247 showing normalized ChIP-seq signal for HSF-1 and Pol II under NHS and HS conditions (ChIP-seq data obtained from Li et al., 2016). HelitronY4_CE sequence containing 96 Hac-HSEs is located upstream of nhr-247 in the wild-type reference strain N2, but is absent in the wild isolate QX1211. (B) Genome browser screenshot depicting the region surrounding the Hac-HSE HSR gene str-96 showing normalized ChIP-seq signal for HSF-1 and Pol II under NHS and HS conditions. HelitronY1A_CE sequence containing 30 Hac-HSEs is located upstream of str-96 in the wild-type reference strain N2, but is absent in the wild isolate QX1211. (C) Genome browser screenshot depicting the region surrounding the Hac-HSE HSR gene fbxa-102 showing normalized ChIP-seq signal for HSF-1 and Pol II under NHS and HS conditions. Helitron2_CE sequence containing nine Hac-HSEs is located upstream of fbxa-102 in the wild-type reference strain N2, but is absent in the wild isolate MY16. (D) Scatterplots showing relative mRNA expression levels (-ΔCt) determined using RT-qPCR (gray open circles) of nhr-247 and str-96 before (NHS) and after heat shock (HS) in N2 worms that have and QX1211 worms that lack upstream Hac-HSEs. Expression levels of the Hin-HSE HSR gene hsp-70 and Helitron1_CE transposase are also shown. RT-qPCR data are normalized to the heat-shock-unchanging housekeeping gene tba-2. Data obtained from three independent biological replicates are shown, with their mean values represented using solid red lines. (E) Scatterplots showing relative mRNA expression levels (-ΔCt) determined using RT-qPCR (gray open circles) of fbxa-102 before (NHS) and after heat shock (HS) in N2 worms that have and MY16 worms that lack upstream Hac-HSEs. Expression levels of the Hin-HSE HSR gene hsp-70 and Helitron1_CE transposase are also shown. RT-qPCR data are normalized to the heat-shock-unchanging housekeeping gene tba-2. Data obtained from three independent biological replicates are shown, with their mean values represented using solid red lines. In N2 NHS worms, fbxa-102 was undetectable in one biological replicate. For panels D-E, a single asterisk represents p<0.05, double asterisks represent p<0.01, and triple asterisks represent p<0.001 (Welch Two Sample t-test). For all genes with the exception of Helitron1_CE transposase, one primer in each qPCR primer pair spans an exon-exon junction to ensure detection of mature mRNA levels.