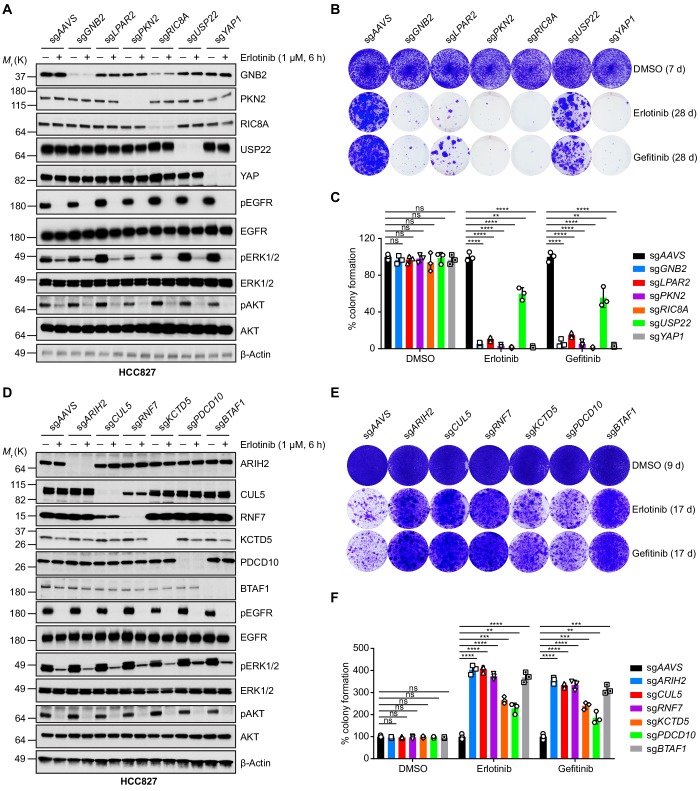
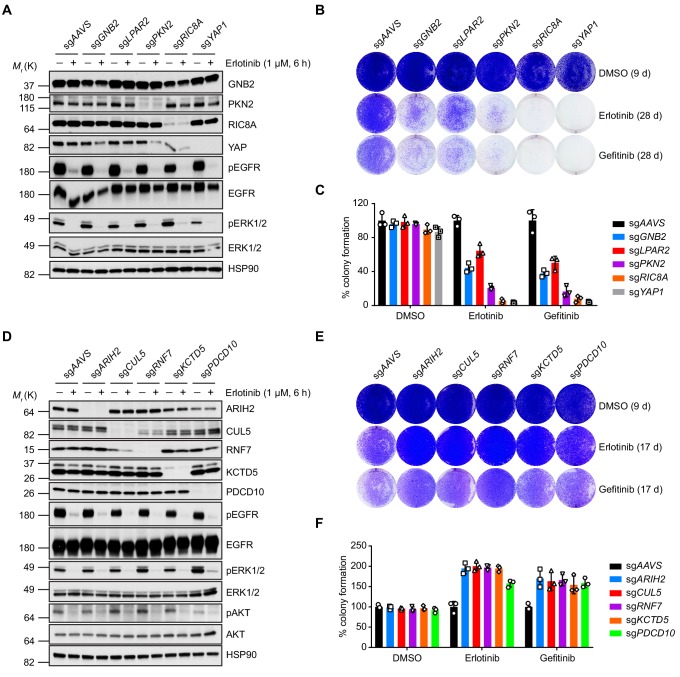
Figure 2. Validation of selected hits by individual knockout in HCC827 cells.
(A) Immunoblots of indicated proteins in cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of negatively selected hits and on-target inhibition of EGFR pathway by erlotinib treatment. β-Actin was used as a loading control. Individual knockout cell lines were generated by lentivirus-mediated expression of sgRNA targeting indicated genes in HCC827 cells with constitutive Cas9 expression. (B) Crystal violet staining colony formation assay of indicated HCC827 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (C) Quantification of colony formation in (B), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± standard deviation (SD) is shown. (D) Immunoblots of indicated proteins in cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of positively selected hits and on-target inhibition of EGFR pathway by erlotinib treatment. β-Actin was used as a loading control. (E) Crystal violet staining colony formation assay of indicated HCC827 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (F) Quantification of colony formation in (E), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± SD is shown. Statistical significance was tested using unpaired two-tailed t test (C and F); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.