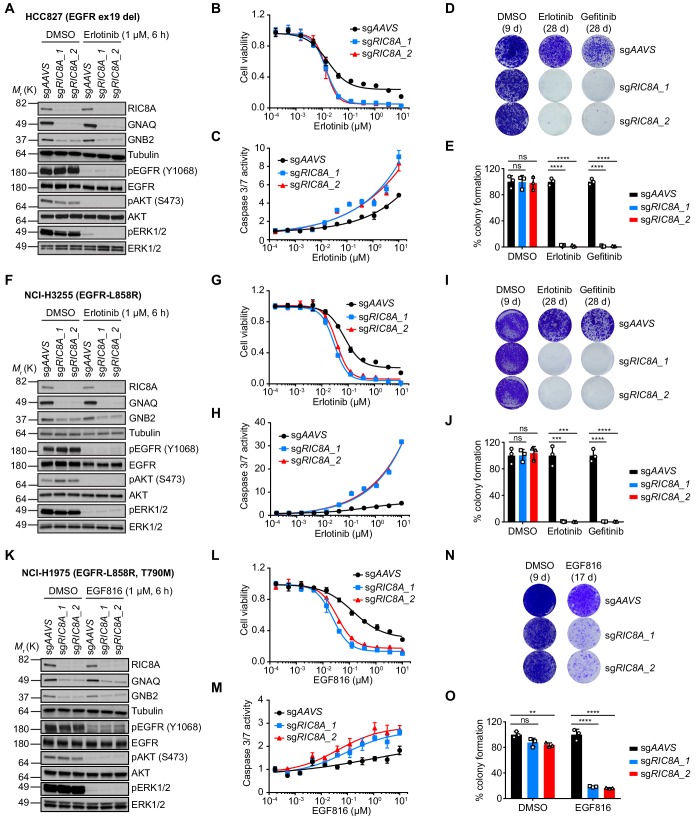
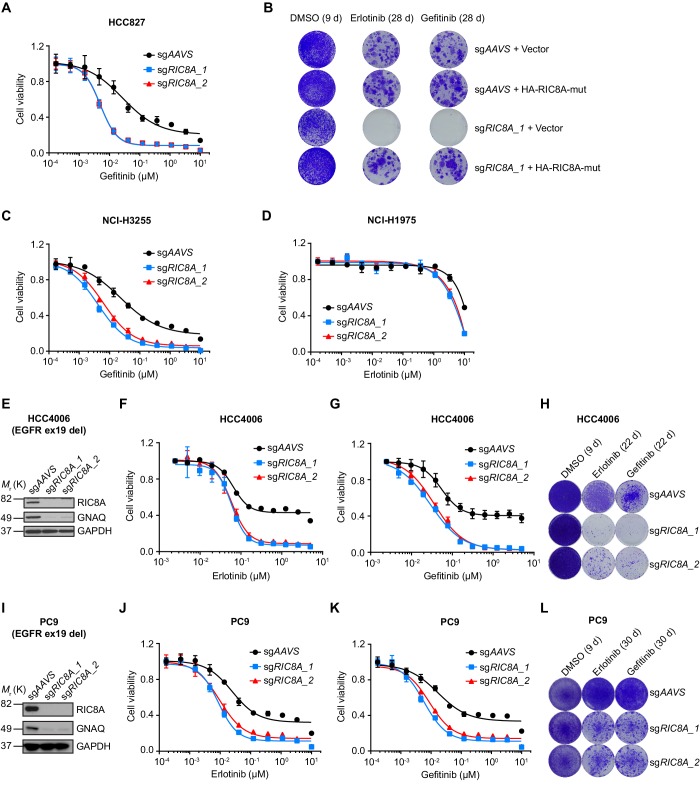
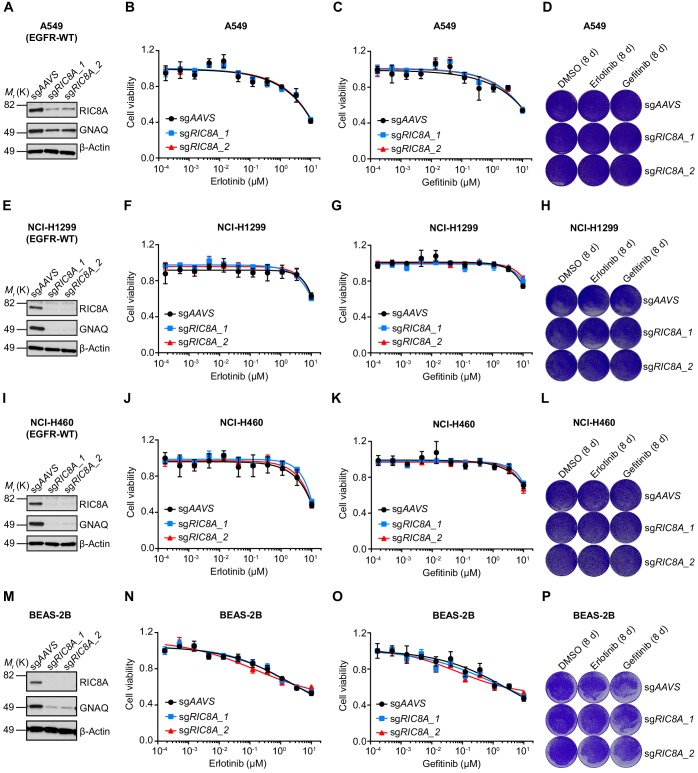
Figure 3. RIC8A depletion causes synthetic lethality with EGFR-TKI in EGFR-mutant NSCLC.
(A) Immunoblots of indicated proteins in control (sgAAVS) or RIC8A knockout (sgRIC8A) HCC827 cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of RIC8A and on-target inhibition of EGFR pathway by erlotinib treatment. Tubulin was used as a loading control. (B) Cell viability assessment by CellTiter-Glo assay of HCC827 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± standard deviation (SD); n = 4. (C) Activated caspase 3/7 measurement of HCC827 cells treated with serial dilutions of erlotinib for 24 hr. Error bars represent mean ± SD; n = 4. (D) Crystal violet staining colony formation assay of indicated HCC827 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (E) Quantification of colony formation in (D), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± SD is shown. (F) Immunoblots of indicated proteins in control or RIC8A knockout NCI-H3255 cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of RIC8A and on-target inhibition of EGFR pathway by erlotinib treatment. Tubulin was used as a loading control. (G) Cell viability assessment by CellTiter-Glo assay of NCI-H3255 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (H) Activated caspase 3/7 measurement of NCI-H3255 cells treated with serial dilutions of erlotinib for 24 hr. Error bars represent mean ± SD; n = 4. (I) Crystal violet staining colony formation assay of indicated NCI-H3255 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (J) Quantification of colony formation in (I), shown as percentage of the sgAAVS sample. Mean (three biological replicates)± SD is shown. (K) Immunoblots of indicated proteins in control or RIC8A knockout NCI-H1975 cells treated with DMSO or EGF816 (1 µM) for 6 hr to confirm specific knockout of RIC8A and on-target inhibition of EGFR pathway by EGF816 treatment. Tubulin was used as a loading control. (L) Cell viability assessment by CellTiter-Glo assay of NCI-H1975 cells treated with serial dilutions of EGF816 for 72 hr. Error bars represent mean ± SD; n = 4. (M) Activated caspase 3/7 measurement of NCI-H1975 cells treated with serial dilutions of EGF816 for 24 hr. Error bars represent mean ± SD; n = 4. (N) Crystal violet staining colony formation assay of indicated NCI-H1975 cell lines treated with DMSO or EGF816 (1 µM). (O) Quantification of colony formation in (N), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± SD is shown. Statistical significance was tested using unpaired two-tailed t test (E, J and O); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.