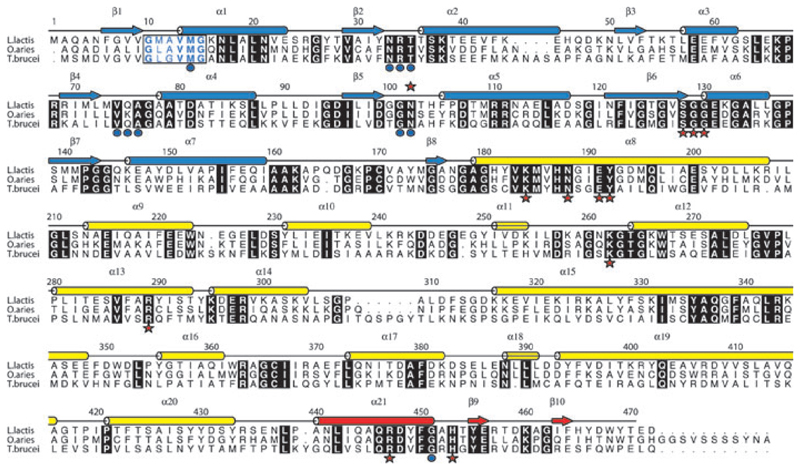
Fig. 2.
Amino acid sequence and secondary structure of LlPDH. Arrows depict β strands, cylinders depict α helices and these are labelled β1–β10 and α1–α21. The elements of secondary structure are coloured according to the domain in which they occur; blue for domain I, yellow for domain II and red for domain III. Aligned sequences of OaPDH and TbPDH are also shown. Most of the amino acids conserved in all three sequences are shown as white letters in black boxes. The exception is the NADP+ fingerprint region (residues 10–15 in LlPDH) where the letters are coloured blue and bold indicates conservation. Red stars identify active site residues that form direct hydrogen bonding interaction with ligands, blue dots identify those residues that interact with cofactor.