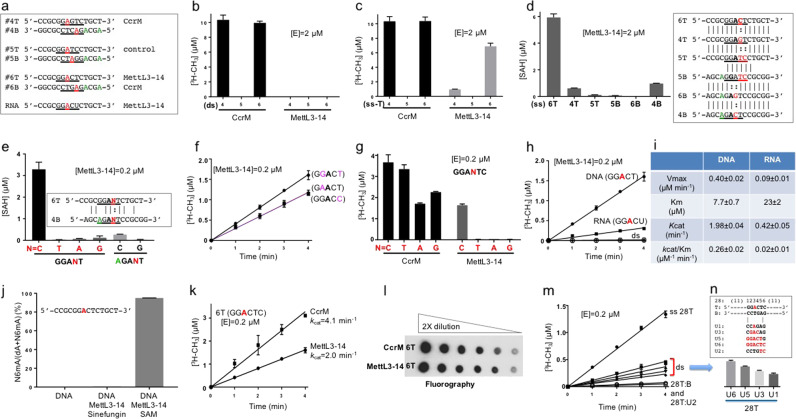
Fig. 1. Human MettL3-14 complex is active on ssDNA and mismatched DNA adenine.
a Olignonuceotides (14-mer) used as substrates. b MettL3-14 is not active on dsDNA. The enzyme concentration [E] is noted in each panel. c, d MettL3-14 is active on ssDNA by two independent assays: incorporation of tritium from 3H-SAM into DNA substrate (panel c) and formation of byproduct SAH in a bioluminescence assay (panel d). Inserted box listed ssDNA sequence alignment. e Substitution of conserved cytosine within GGACT diminished Mettl3-14 activity. f Replacement of the second guanine-to-adenine (GAACT, red) or the last thymine-to-cytosine (GGACC, blue) retained comparable activity. The two lines fell on top to each other, giving rise to purple. g Comparison of CcrM (GAnTC) and MettL3-14 (GGACT) on ssDNA oligo 6 T and its derivatives. h Comparison of MettL3-14 on oligo 6 T and its corresponding RNA. i Comparison of the kinetic parameters on ssDNA and ssRNA, derived from Supplementary Fig. S2d. j Quantitative measurement of N6mA by mass spectrometry, derived from Supplementary Fig. 3. k, l Comparison of CcrM and MettL3-14 on oligo 6 T. m, n MettL3-14 is active on 28-bp dsDNA-containing mismatched pairs. Data represent the mean ± SD of three independent determinations (N = 3) performed in duplicate.