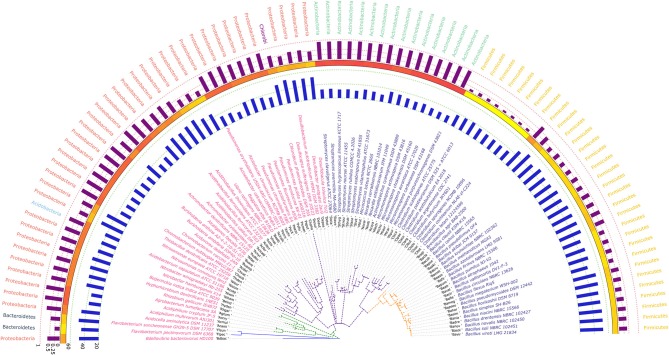
Figure 4.
A phylogenetic tree showing the relationship between the soil bacterial species considered in this study based on 16S rRNA gene sequences along with Gram nature, taxonomic position and codon usage annotation data. The name of the species have been depicted in color corresponding to its Gram nature with magenta and blue representing Gram negative and positive nature, respectively. The outermost semicircle with magenta bars represents the genomic GC3 while the innermost semicircle with blue bars represents the genomic Nc. The middle strip with yellow to red color gradient depicts the genomic GC content with red representing maximum GC content. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model (Kimura, 1980). The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The tree with the highest log likelihood (−6,331.0306) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0.5623)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 40.6240% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 357 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Kumar et al., 2008). The visualization and annotation of the phylogenetic tree was done using iTOL ver. 4.4.2 (Letunic and Bork, 2007).