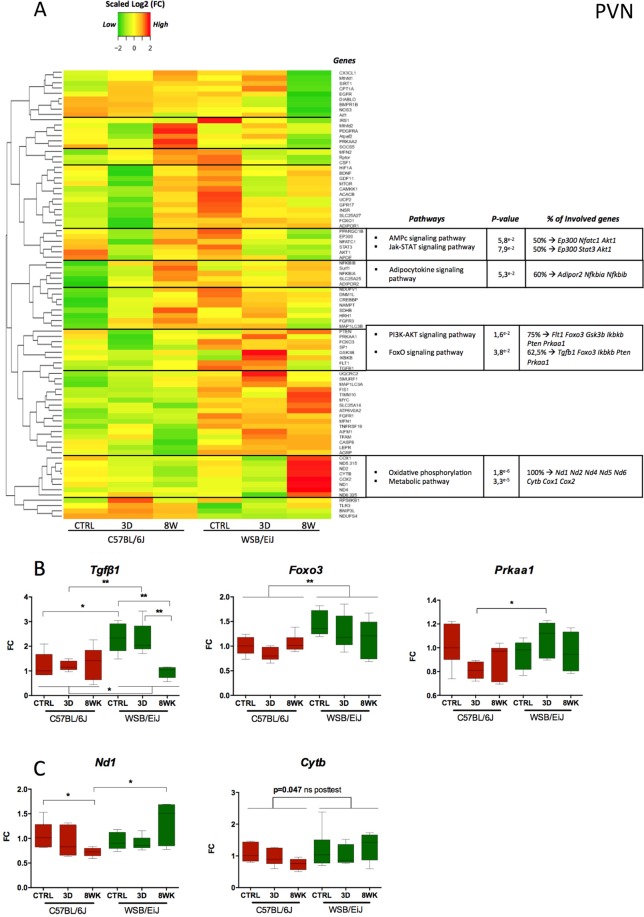
Figure 9.
Expression profiles of genes related to inflammation or mitochondria are different between both strains, and also differentially influenced by HFD in the PVN. (A) Heatmap showing for the paraventricular nucleus of the hypothalamus (PVN), the relative gene expression data (expressed as scaled log2 Fold Change values) for 86 genes related to inflammatory and mitochondrial pathways: Data were collected by microfluidic qRT-PCR from PVN of twelve week-old C57BL/6J (left panel) and WSB/EiJ (right panel) mice either maintained under control feeding (CTRL), or challenged with HFD for the last 3 days of the period (3D) or during the whole 8-week experiment duration (8WK). Color transition represents gene expression levels expressed as scaled log2 Fold Change values (scaled Log2(FC)): green for low expression, yellow for mean expression and red for high expression. Genes with similar expression profiles were clustered together with “Heatmap2” R script and the cluster delimited from the dendrogram by a black horizontal line. Each cluster of genes was analysed against the 86 genes with DAVID bioinformatics tools and significantly enriched pathways were identified. The summary of the pathways is shown in the table (at the right of each cluster) with the associated significant P-value (P ≤ 0.1), the percentage of genes in the cluster involved in each pathway, and the names of the involved genes. (B) Gene expression data for representative significantly differentially regulated genes involved in pathways defined in (A): Data were collected by microfluidic qRT-PCR from microdissected PVN of twelve week-old C57BL/6J (red) and WSB/EiJ (green) mice either maintained under control feeding (CTRL), or challenged with HFD for the last 3 days of the period (3D) or during the whole 8-week experiment duration (8WK). Tgfβ: Transforming growth factor beta Transcription factor; Foxo3: forkhead box O-3; Prkka1: Protein kinase AMP-activated catalytic subunit alpha 1; Nd1: NADH-ubiquinone oxidoreductase chain 1; Cytb: Cytochrome b. Non parametric Two-way Anova analysis with permutations revealed a significant combined effect of strain and diet on Tgfβ and Nd1 expressions, whereas Foxo3 expression is significantly different in the two strains independently of the diet, and Prkka1 expression is regulated by the diet, independently of the strain. Post Hoc tests results are indicated on the graph. (*, P ≤ 0.05; **, P ≤ 0.01. N = 5–7 per group). FC: relative fold-change expression compared to C57BL/6J control group.