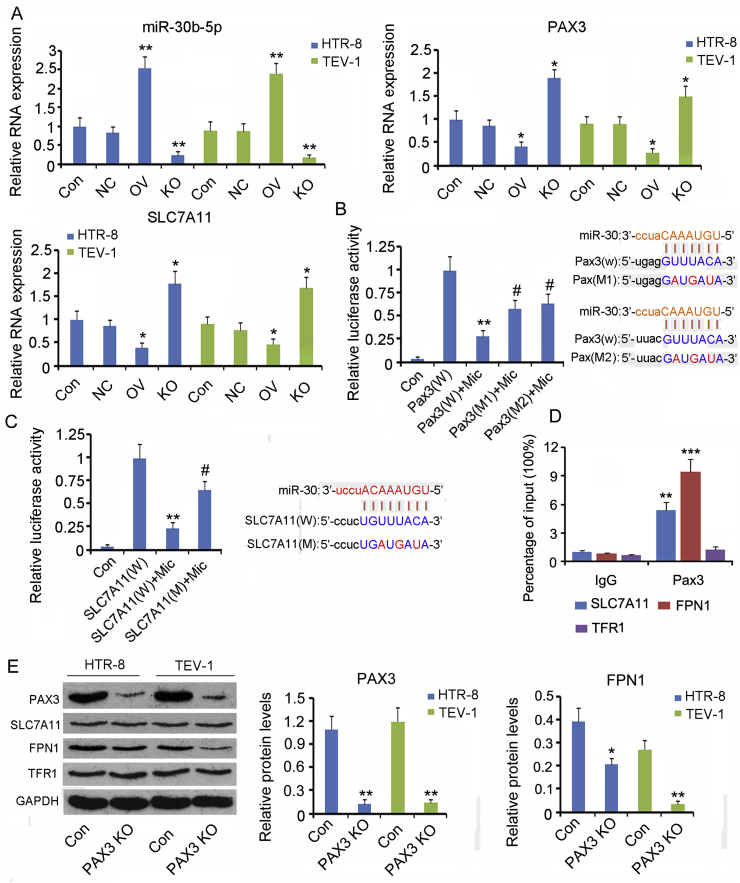
Fig. 6.
The regulatory effects of miR-30b-5p and Pax3 on gene expression.
(A) Artificial upregulation and downregulation of miR-30b-5p expression using mimics and inhibitors, respectively. Expression of miR-30b-5p, Pax3, and SLC7A11 in cells was assessed using PCR. NC: transfection of negative control of miRNA vectors; OV: miR-30b-5p overexpression; KO: miR-30b-5p knockdown. *p < 0.05, **p < 0.01, compared with the control group. (B and C) Luciferase reporter assay was performed to identify the putative binding sites (seed sequences) for hsa-miR-30b-5p in the 3′-UTR of pax3 and SLC7A11 mRNA, as predicted by the bioinformatic analysis webs, including TargetScan, miRanda, and PicTar. The wild-type (WT) pax3 and SLC7A11 3′-UTR, as well as their mutant versions (MT), with mutations in the putative binding sites, were synthesized and inserted into a pmirGLO vector at XhoI and NotI restriction sites. HTR-8/SVneo cells were transfected with the WT or MT constructs, along with either hsa-miR-10b-3p mimics or negative control. The activity of firefly luciferase was measured and normalized to that of Renilla luciferase. Pax3(W): wild-type pax3 constructs; Pax3(M1) and Pax3(M2): two mutant type of Pax3 constructs; SLC7A11(W) and SLC7A11 (M): wild-type and mutant type of Pax3 constructs, respectively; Mimics: hsa-miR-10b-3p mimics. *p < 0.05, **p < 0.01, compared with the Pax3(W) and SLC7A11(W) groups; #p < 0.05, ##p < 0.01, compared with the Pax3(W) + Mimics and SLC7A11(W) + Mimics groups. (D) ChIP assay was performed to determine whether Pax3 could bind to the promoters of SLC7A11, FPN1 and TFR1 genes. **p < 0.01, ***p < 0.001, compared with the IgG group. (E) Pax3 in HTR-8/SVneo and TEV-1 cells was knocked down, followed by the evaluation of protein levels of SLC7A11, FPN1, and TFR1 using western blot assay. Pax3 KO: Pax3 knockdown. *p < 0.05, **p < 0.01, compared with the control group.