Abstract
Myeloid/lymphoid neoplasms with PDGFRB rearrangement are a distinct type of myeloid neoplasms that occur in association with rearrangement of PDGFRB at 5q32. The hematological features most often show prominent eosinophilia. We herein report a patient with myeloid/lymphoid neoplasms with PDGFRB rearrangement with t(5;10)(q33;q22) who showed atypical chronic myeloid leukemia-like clinical features without eosinophilia and achieved an optimal response to imatinib. A sequence analysis showed a CCDC6-PDGFRB fusion gene with a new break point in the PDGFRB gene. This is the sixth case of myeloid/lymphoid neoplasm with PDGFRB rearrangement harboring a CCDC6-PDGFRB fusion gene, and it has a new breakpoint in the PDGFRB fusion gene.
Keywords: myeloid/lymphoid neoplasms with PDGFRB rearrangement, atypical chronic myeloid leukemia, t(5;10)(q33;q22), CCDC6-PDGFRB, imatinib
Introduction
Myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB or FGFR1 were defined in the World Health Organization (WHO) classification in 2008 as a distinct clinical entity with characteristic gene rearrangements (1). Later, PCM1-JAK2 rearrangement was added to the category in WHO classification 2016 as a provisional entity (2-5). Within this category, some features are shared, while others differ, but all of the neoplasms are induced by the formation of a fusion gene, resulting in the expression of an aberrant tyrosine kinase and making the disease responsive to tyrosine kinase inhibitors. The disease category is now distinguished from atypical chronic myeloid leukemia (aCML), BCR-ABL1-negative, the neoplastic cells of which do not have PDGFRA, PDGFRB, FGFR1 or PCM1-JAK2 (2,5). Eosinophilia is characteristic but not crucial for the diagnosis (2,6).
PDGFRA-related disorders usually present as chronic eosinophilic leukemia with prominent involvement of the mast cell lineage and sometimes the neutrophil lineage. The most common PDGFRA rearrangement is FIP1L1-PDGFRA (7,8). In the setting of PDGFRB-related disease, the features of myeloproliferative neoplasm (MPN) are more variable but are often similar to those of chronic myelomonocytic leukemia (CMMoL) with eosinophilia. Myeloid/lymphoid neoplasm with PDGFRB rearrangement occurs in association with rearrangement of PDGFRB at 5q32. Among these variants, t(5;12)(q32;p13.2) with the formation of an ETV6-PDGFRB fusion gene is the most common, and imatinib has been reported to be effective (9-12). In uncommon variants, other translocations with a 5q32 breakpoint lead to the formation of other fusion genes, also incorporating part of PDGFRB. Translocation with chromosome 10 is rare, and only five cases with t(5;10) translocation have been reported thus far (13-17). The translocation t(5;10) causes the fusion of the PDGFRB tyrosine kinase gene at the 5q33 region with CCDC6 on chromosome 10q21.2 (13-18). These five cases had the same breakpoints of the CCDC6-PDGFRB fusion gene, but most were reported as aCML with eosinophilia before 2008 (5).
We herein report a patient with myeloid/lymphoid neoplasms with PDGFRB rearrangement with chromosomal reciprocal translocation t(5;10)(q33;q22) who showed aCML-like clinical features without eosinophilia and achieved an optimal response to imatinib. A sequence analysis showed the CCDC6-PDGFRB fusion gene with a new break point.
Case Report
A 53-year-old man was referred to our hospital due to leukocytosis. He had a medical history of hypertension without any family history. The patient had no symptoms, and his Eastern Cooperation Oncology Group (ECOG) performance status score was zero.
A physical examination revealed splenomegaly (3 cm below the costal margin). However, hepatomegaly was not observed. Complete blood count examination results showed the following: leukocyte count: 64.2×109/L (blasts: 0.5%, promyelocytes: 1.0%, myelocytes: 2.5%, metamyelocytes: 6.0%, stab neutrophils: 7.5%, segmented neutrophils: 77.5%, eosinophils: 0.5%, basophils: 0.5%, monocytes: 1.5%, and lymphocytes: 2.5%), red blood cell count: 4.67×109/L, hemoglobin level: 13.5 g/dL, and platelet count: 290×109/L. Laboratory data showed that lactate dehydrogenase level increased to 375 U/I. Bone marrow (BM) aspiration results showed marked myeloid hypercellular marrow with a nucleated cell count of 80.5×104/μL and high myeloid/erythroid ratio of 7.9. In addition, 1% myeloblasts and 9% promyelocytes were observed, indicating CML in the chronic phase. A flow cytometric analysis did not show any specific cell population. However, a chromosomal analysis of the BM showed that a majority of metaphases had reciprocal translocation of t(5;10)(q33;q22) in all cells that were analyzed [20/20]. Major and minor BCR-ABL1 fusion genes were not detected via real-time polymerase chain reaction (RT-PCR). JAK2 V617F mutation was also negative. A fluorescence in situ hybridization (FISH) analysis using 3' and 5' probes of the PDGFRB gene showed that PDGFRβ rearrangement was positive in 95% of granulocytes in BM cells and in 95% of those in peripheral blood cells. In contrast, cutaneous fibroblasts had no PDGFRβ rearrangement. These data suggested that the patient had a myeloid/lymphoid neoplasm with PDGFRB gene rearrangement.
Materials and methods
To evaluate the molecular response, we identified the counterpart gene to the PDGFRB gene and its breakpoint. BM cells, peripheral blood cells, and cutaneous fibroblasts were obtained after the patient provided their informed consent.
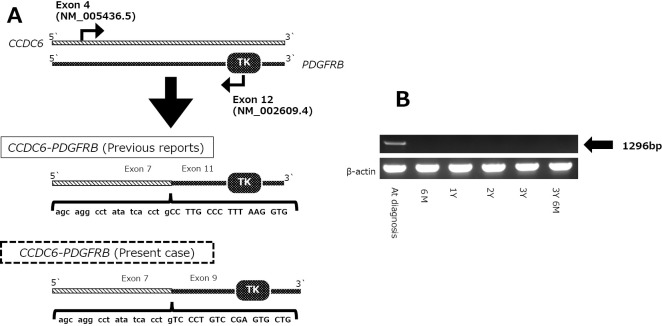
Mononuclear cells were separated from the BM and peripheral blood via density gradient centrifugation using Ficoll-Paque PLUS (General Electronics Healthcare, Chicago, USA). Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany), and cDNA was generated by the reverse transcription of 200 ng RNA with PrimeScriptTM RT Reagent Kit (TaKaRa Bio, Shiga, Japan). The PCR protocol was as follows: 35 cycles for 30 seconds at 95℃, 30 seconds at 60℃, and 2 minutes at 74℃, with an initial 5 minutes at 95℃. Previous studies showed that CCDC6 is fused at exon 7 (NM_005436.5) in frame to the PDGFRB gene beginning at exon 11 (NM_002609.4), so we selected sense and antisense primers directed to CCDC6 exon 4 and PDGFRB exon 12, respectively (Figure A) (18). The primers were as follows: sense primer, 5'-AATCGCCTCTGGAAAAGGAT-3'; antisense primer, 5'-CCTTCCATCGGATCTCGTAA-3'. The PCR products were electrophoresed on 1.5% agarose gels.
Figure.
Detection of the CCDC6-PDGFRB fusion gene and identification of its breakpoint and effect of imatinib treatment. (A) Schematic illustration of real-time polymerase chain reaction (PCR) and DNA sequencing. To detect the CCDC6-PDGFRB fusion gene, sense and antisense primers directed at CCDC6 exon 4 (NM_005436.5) and PDGFRB exon 12 (NM_002609.4), respectively, were used. By sequencing the PCR products [PCR band at the diagnosis in (B)], the novel breakpoint was identified. (B) Real-time PCR (RT-PCR) of the bone marrow samples at the diagnosis and at several time points after initiating imatinib. Six months after starting imatinib treatment, the RT-PCR results were negative and have remained the same since then.
The PCR products were subsequently subjected to direct sequencing with Big Dye terminator V 3.1 cycle sequencing reagents (Applied Biosystems, Waltham, USA) according to the manufacturer's protocol. For breakpoint identification, the samples were analyzed on an ABI PRISM 3,100 Genetic Analyzer (Applied Biosystems). Molecular monitoring of minimal residual disease was performed via RT-PCR.
Results
Detection of CCDC6-PDGFRB fusion gene and its breakpoint
CCDC6-PDGFRB chimeric mRNA was detected in the patient's RNA but not in the control RNA. The cDNA sequence was then compared with sequences in the National Center for Biotechnology Information (NCBI) RefSeq. We confirmed that CCDC6 exon 7 is fused to PDGFRB exon 9. This suggests that the breakpoint in CCDC6 occurred between exons 7 and 8 and resulted in in-frame fusion to PDGFRB. It also suggests that the breakpoint in PDGFRB was located between exons 8 and 9 (Figure B).
Clinical course
Based on the presence of PDGFRB rearrangement, treatment with imatinib at a daily dose of 400 mg was initiated. The therapy was well-tolerated, and serious adverse effects were not observed. A complete cytogenetic response to imatinib was achieved after three months of therapy as confirmed by chromosome and FISH analyses. An RT-PCR analysis showed no band at six months, indicating molecular remission. The patient has remained RT-PCR-negative for three years with imatinib treatment.
Discussion
In this report, we identified a novel breakpoint in the CCDC6-PDGFRB fusion gene in a patient with myeloid/lymphoid neoplasm with PDGFRB gene rearrangement with t(5;10)(q33;q22). This enabled the monitoring of the molecular response, and the patient became RT-PCR-negative after six months of imatinib treatment and has continued with such treatment for three years.
Five cases involving the translocation of chromosomes 5 and 10 have been reported thus far (13-18). In WHO classification 2008, aCML was defined as a leukemic disorder with myelodysplastic as well as myeloproliferative features without BCR-ABL1, PDGFRA, PDGFRB, FGFR1 or PCM1-JAK2 rearrangements (1). However, before 2008, patients with myeloproliferative features having these gene rearrangements were diagnosed with aCML (19,20). Therefore, four of these cases were reported as aCML with eosinophilia, and the other was reported as CMMoL (Table). Two cases had t(5;10)(q33;q21), and three had t(5;10)(q33;q22). The patients were all middle-aged men with mild eosinophilia, BM fibrosis, splenomegaly, and bone pain. A Southern blot analysis with PDGFRB cDNA probe was performed, and the rearranged bands were detected in all cases. A sequencing analysis showed that these five cases had the same breakpoint of exon 7 at the CCDC6 gene and exon 11 at the PDGFRB gene.
Table.
Characteristics of Patients with T(5;10) Chromosomal Abnormality.
| No. | Diagnosis | Age (years)/ sex |
WBC count (×109/L) | Eos (%) | Immature leukocytes (%) | Cytogenetics | Breakpoint at the CCDC6 | Breakpoint at the PDGFRB | Treatment | Response to imatinib | References (No.) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | aCML | 49/M | 33 | 6 | 45 | t(5;10)(q33;q22) | exon 7 | exon 11 | HU, Ara-C, 6-TG | NA | 13, 18 |
| 2 | aCML | 48/M | 94 | 7 | NA | t(5;10)(q33;q21.2) | exon 7 | exon 11 | HU, Allo-SCT | NA | 14 |
| 3 | aCML | 44/M | 158 | 8 | 13 | t(5;10)(q33;q22) | exon 7 | exon 11 | Imatinib | CCyR; 3 weeks | 15 |
| 4 | aCML | 49/M | 7.4 | 6 | 5 | t(5;10)(q33;q21) | exon 7 | exon 11 | Imatinib | Failure; 12 weeks | 17 |
| 5 | CMMoL | NA | NA | NA | NA | t(5;10)(q33;q22) | exon 7 | exon 11 | Imatinib | CCyR; 24 months | 16 |
| 6 | aCML | 53/M | 64 | 0.5 | 10 | t(5;10)(q33;q22) | exon 7 | exon 9 | Imatinib | CCyR; 6 months | Present case |
aCML: atypical chronic myeloid leukemia, CMMoL: chronic myelomonocytic leukemia, M: male, WBC: white blood cell. Eos: eosinophil, HU: hydroxyurea, 6-TG: 6-thioguanine, Allo-SCT: allogeneic stem cell transplantation, CCyR: complete cytogenetic response, NA: not applicable
However, a sequencing analysis of the PCR products of the CCDC6-PDGFRB fusion gene in our patient showed that CCDC6 exon 7 was fused to PDGFRB exon 9. The breakpoint in CCDC6 was the same as that in the previously reported cases. However, the breakpoint in PDGFRB was different.
The CCDC6 (also known as H4 or D10S170) gene encodes a 585-amino acid protein with no significant homology to known genes and an unknown function (13,18). CCDC6 rearrangement is observed in approximately 20% of human papillary carcinomas (18). In vitro studies have shown that the leucine zipper region of CCDC6 included in the fusion is responsible for the dimerization of the PTC1 oncoprotein and is essential for tyrosine hyperphosphorylation and transformation in vitro. Of the myeloid/lymphoid neoplasm with PDGFRB rearrangements, ETV6-PDGFRB is the most common (21). Since the number of cases reported to have the CCDC6-PDGFRB fusion gene is extremely small, differences from cases with other fusion genes with PDGFRB have not been identified. Compared to the previously reported cases with the CCDC6-PDGFRB fusion gene, eosinophilia or granulocytic dysplasia and BM fibrosis were not observed in our patient. Whether or not this could be attributed to the differences in the breakpoint of the CCDC6-PDGFRB fusion gene was not clear.
The PDGFRB fusion gene encodes constitutively activated receptor tyrosine kinases that can be inhibited by imatinib. David et al. have shown durable hematologic and cytogenetic responses that were achieved with imatinib in a patient with BCR-ABL-negative chronic myeloproliferative disorder with the PDGFRB fusion gene (11). Three patients were treated with imatinib at a daily dose of 400 mg (15-17). Two achieved cytogenetic response after 3 weeks and 24 weeks of imatinib therapy (15,16). One patient did not respond to imatinib and could not continue the treatment due to severe hematologic toxicity (17). Imatinib treatment was initiated three years from the onset. However, myelofibrosis was already observed in the BM. These findings suggested that starting imatinib therapy at an early stage is important for achieving a good response to imatinib. In the present case, the patient had a good response to imatinib. Differences in the breakpoint did not influence the effect of imatinib treatment.
To our knowledge, this is the sixth case of myeloid/lymphoid neoplasm with PDGFRB rearrangement harboring a CCDC6-PDGFRB fusion gene. Of note, a sequencing analysis revealed a novel breakpoint; however, whether or not the difference in the breakpoint is associated with clinical characteristics, such as dysplasia, eosinophilia, and fibrosis in the BM, is unclear at present. Further studies must be conducted in order to better understand myeloid/lymphoid neoplasm with PDGFRB rearrangement.
The authors state that they have no Conflict of Interest (COI).
References
- 1. Vardiman JW, Thiele J, Arber DA, et al. . The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 114: 937-951, 2009. [DOI] [PubMed] [Google Scholar]
- 2. Arber DA, Orazi A, Hasserjian R, et al. . The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127: 2391-2405, 2016. [DOI] [PubMed] [Google Scholar]
- 3. Bousquet M, Quelen C, De Mas V, et al. . The t(8;9)(p22;p24) translocation in atypical chronic myeloid leukaemia yields a new PCM1-JAK2 fusion gene. Oncogene 24: 7248-7252, 2005. [DOI] [PubMed] [Google Scholar]
- 4. Reiter A, Walz C, Watmore A, et al. . The t(8;9)(p22;p24) is a recurrent abnormality in chronic and acute leukemia that fuses PCM1 to JAK2. Cancer Res 65: 2662-2667, 2005. [DOI] [PubMed] [Google Scholar]
- 5. Swerdlow SH, Campo E, Harris NL, et al. . WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th ed. World Health Organization, 2017. [Google Scholar]
- 6. Gotlib J. World Health Organization-defined eosinophilic disorders: 2017 update on diagnosis, risk stratification, and management. Am J Hematol 92: 1243-1259, 2017. [DOI] [PubMed] [Google Scholar]
- 7. Cools J, DeAngelo DJ, Gotlib J, et al. . A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med 348: 1201-1214, 2003. [DOI] [PubMed] [Google Scholar]
- 8. Gotlib J, Cools J, Malone JM 3rd, Schrier SL, Gilliland DG, Coutre SE. The FIP1L1-PDGFRalpha fusion tyrosine kinase in hypereosinophilic syndrome and chronic eosinophilic leukemia: implications for diagnosis, classification, and management. Blood 103: 2879-2891, 2004. [DOI] [PubMed] [Google Scholar]
- 9. Apperley JF, Gardembas M, Melo JV, et al. . Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med 347: 481-487, 2002. [DOI] [PubMed] [Google Scholar]
- 10. Reiter A, Gotlib J. Myeloid neoplasms with eosinophilia. Blood 129: 704-714, 2017. [DOI] [PubMed] [Google Scholar]
- 11. David M, Cross NC, Burgstaller S, et al. . Durable responses to imatinib in patients with PDGFRB fusion gene-positive and BCR-ABL-negative chronic myeloproliferative disorders. Blood 109: 61-64, 2007. [DOI] [PubMed] [Google Scholar]
- 12. Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77: 307-316, 1994. [DOI] [PubMed] [Google Scholar]
- 13. Siena S, Sammarelli G, Grimoldi MG, et al. . New reciprocal translocation t(5;10)(q33;q22) associated with atypical chronic myeloid leukemia. Haematologica 84: 369-372, 1999. [PubMed] [Google Scholar]
- 14. Kulkarni S, Heath C, Parker S, et al. . Fusion of H4/D10S170 to the platelet-derived growth factor receptor beta in BCR-ABL-negative myeloproliferative disorders with a t(5;10)(q33;q21). Cancer Res 60: 3592-3598, 2000. [PubMed] [Google Scholar]
- 15. Garcia JL, Font de Mora J, Hernandez JM, et al. . Imatinib mesylate elicits positive clinical response in atypical chronic myeloid leukemia involving the platelet-derived growth factor receptor beta. Blood 102: 2699-2700, 2003. [DOI] [PubMed] [Google Scholar]
- 16. Drechsler M, Hildebrandt B, Kundgen A, Germing U, Royer-Pokora B. Fusion of H4/D10S170 to PDGFRbeta in a patient with chronic myelomonocytic leukemia and long-term responsiveness to imatinib. Ann Hematol 86: 353-354, 2007. [DOI] [PubMed] [Google Scholar]
- 17. Bastie JN, Garcia I, Terre C, Cross NC, Mahon FX, Castaigne S. Lack of response to imatinib mesylate in a patient with accelerated phase myeloproliferative disorder with rearrangement of the platelet-derived growth factor receptor beta-gene. Haematologica 89: 1263-1264, 2004. [PubMed] [Google Scholar]
- 18. Schwaller J, Anastasiadou E, Cain D, et al. . H4(D10S170), a gene frequently rearranged in papillary thyroid carcinoma, is fused to the platelet-derived growth factor receptor beta gene in atypical chronic myeloid leukemia with t(5;10)(q33;q22). Blood 97: 3910-3918, 2001. [DOI] [PubMed] [Google Scholar]
- 19. Alimena G, Breccia M, Mancini M, et al. . Clonal evolution in Philadelphia chromosome negative cells following successful treatment with Imatinib of a CML patient: clinical and biological features of a myelodysplastic syndrome. Leukemia 18: 361-362, 2004. [DOI] [PubMed] [Google Scholar]
- 20. Breccia M, Biondo F, Latagliata R, Carmosino I, Mandelli F, Alimena G. Identification of risk factors in atypical chronic myeloid leukemia. Haematologica 91: 1566-1568, 2006. [PubMed] [Google Scholar]
- 21. Cross NC, Reiter A. Tyrosine kinase fusion genes in chronic myeloproliferative diseases. Leukemia 16: 1207-1212, 2002. [DOI] [PubMed] [Google Scholar]