Figure 7.
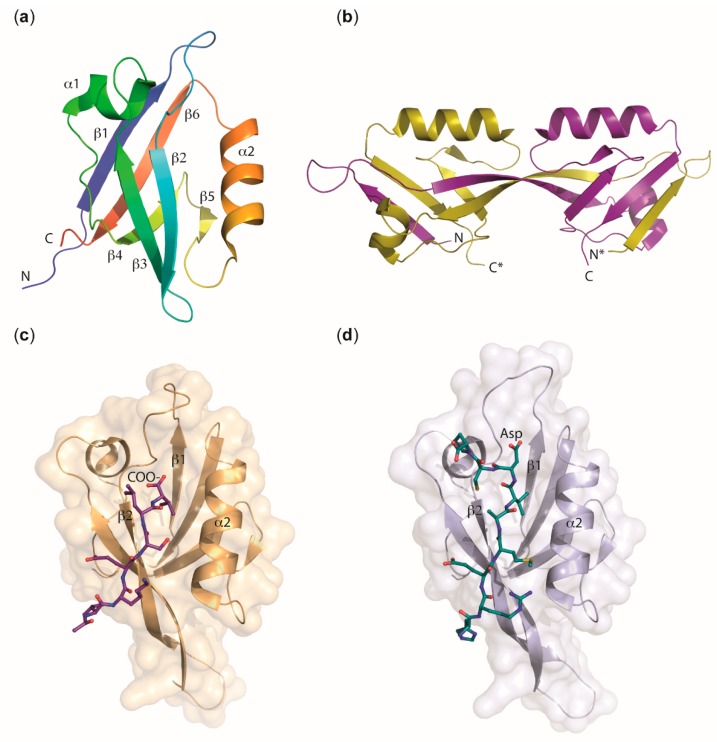
Structural features of PDZ domains. (a) Topology of a prototypical PDZ domain, here PALS 1 PDZ, PDB entry 4UU6 [87]. The polypeptide chain is drawn in rainbow colors changing from N- to C-terminus. A cleft for binding ligand peptides in an extended conformation is visible between strand β2 and helix α2. (b) Domain-swapped dimer formed by ZO-2 PDZ2, PDB entry 3E17 [88]. The two polypeptide chains are drawn in yellow and purple. The chain marked with an asterisk (*) corresponds to the second monomer within the dimer. Domain swapping moves the N-terminal β1 strand and half of β2 of one chain into the core structure of the other, leaving the ligand-binding geometry in both halves of the dimer intact. The PDZ2 domains in ZO-1, ZO-2, and ZO-3 are all found in the domain-swapped dimeric form [80,88,89,90,91]. (c) Canonical binding of a C-terminal ligand peptide to a PDZ domain, here PAR-6 PDZ bound to the hexapeptide VKRSLV, PDB entry 1RZX [92]. The terminal carboxy group is bound to the carboxylate-binding loop between strands β1 and β2 of PAR-6 PDZ and the extended ligand peptide aligns in antiparallel orientation with β2, extending the β-sheet. Note that this binding mode is energetically disfavored for PAR-6 PDZ and requires binding of CDC42 at a nearby CRIB domain (not shown). (d) Binding of an internal peptide to a PDZ domain, here PAR-6 PDZ bound to a dodecapeptide representing amino acids 29–40 of PALS1, PDB entry 1X8S [93]. The ligand peptide adopts an extended conformation with an aspartic acid side chain mimicking the carboxy group of the canonical C-terminal peptide ligand. Note the altered conformation of the carboxylate-binding loop.