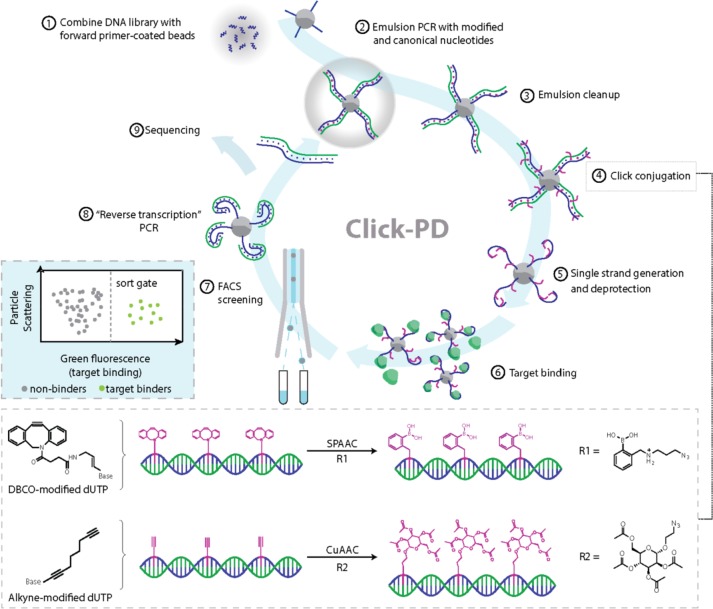
Figure 1.
Click-PD strategy for the synthesis and screening of base-modified aptamers. After combining the initial DNA library with forward primer-coated magnetic beads (step 1), we perform emulsion PCR (step 2) to produce monoclonal aptamer particles in which dT is substituted with alkyne modified-dUTP. We then break the emulsions (step 3) and use either strain-promoted alkyne–azide cycloaddition (SPAAC) or copper(I)-catalyzed alkyne–azide cycloaddition (CuAAC) (step 4; bottom box) to conjugate azide-modified functional groups to the alkyne group on the modified uracil nucleotides. R1= azido-phenylboronic acid, R2 = 2-azidoethyl 2,3,4,6-tetra-O-acetyl-α-d-mannopyranoside. These are converted to single-stranded aptamers (step 5) containing modified deoxyuridine and combined with fluorescently labeled target molecules (step 6). FACS screening allows us to isolate molecules that exhibit high-affinity target binding (step 7, left inset box). The selected base-modified aptamers are then converted back to natural DNA via a reverse transcription-like PCR reaction (step 8) and subjected to sequencing (step 9) or further screening.