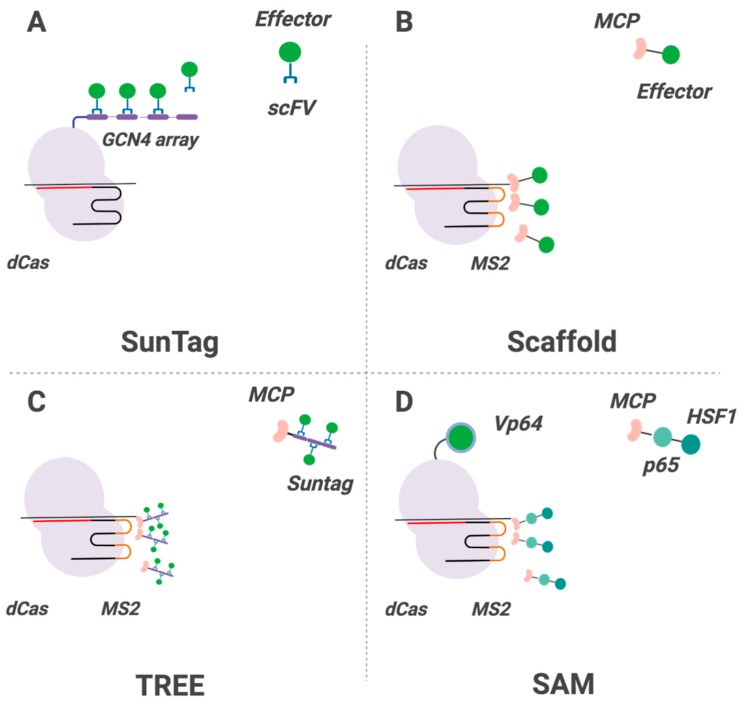
Figure 2.
Modification of CRISPR components for improved epigenetic regulation. (A) SunTag technique. dCas is fused with GCN4 peptide array, which attracts any effector molecule containing single-chain variable fragments (scFV). Multiple GCN4-scFV interactions ensure efficient recruitment of many effector molecules to the dCas-programmed genomic site. (B) Scaffold technique. In Scaffold, effector molecules are recruited to the target site via the interaction of MCP aptamer-specific protein with a short synthetic gRNA containing MS2 aptamer. gRNA protrudes out of the Cas-gRNA complex, so that chimeric gRNA-MS2 transcripts efficiently recruit effectors carrying MCP molecules. (C) TREE combines SunTag and Scaffold techniques, providing additional recruitment of effector molecules. (D) SAM is based on a two-component transcriptional effector (p65-HSF1) recruited to the target via MS2-MCP interaction. Additionally, dCas protein is tethered to a transcriptional regulator (VP64) to increase potency of the effect. This picture was created in BioRender software.