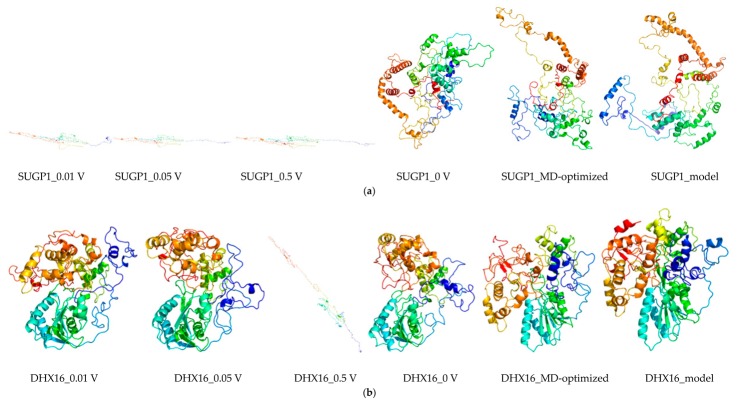
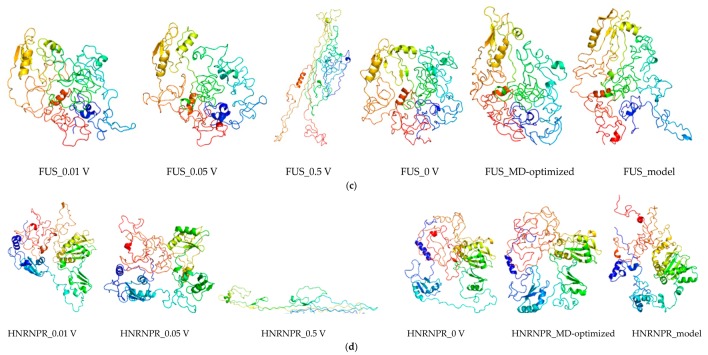
Figure 8.
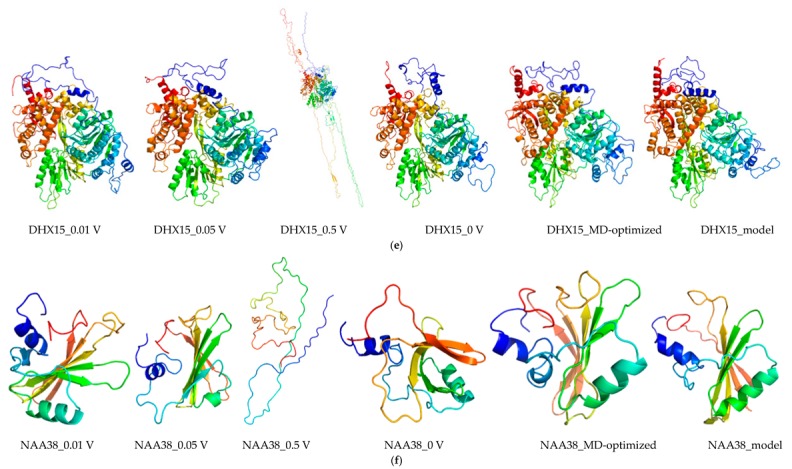
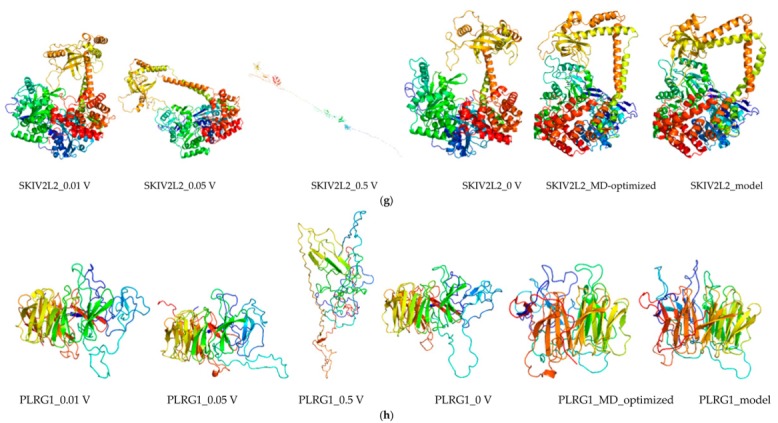
The structure of the 3D protein of hub proteins optimized by molecular dynamics. (a) SUGP1_model: the three-dimensional structure of the SUGP1 protein obtained by modeling; SUGP1_MD-optimized: after at least 100 ns molecular dynamics simulation, the lowest energy protein conformation of SUPG1 protein was obtained (based on the three-dimensional structure of the primary modeling) and was subsequently used for subsequent molecular dynamics simulations. After simulation of different electric field conditions, including 0 V (SUGP1_0 V), 0.01 V (SUGP1_0.01 V), 0.05 V (SUGP1_0.05 V) and 0.5 V (SUGP1_0.5 V), the lowest energy protein of SUGP1 protein were obtained respectively. Other proteins (b–h) were treated similarly to the SUPG1 protein. The pictures were drawn by the Visual Molecular Dynamics (VMD) software and the color map of the protein structure was shown in terms of protein secondary structure.