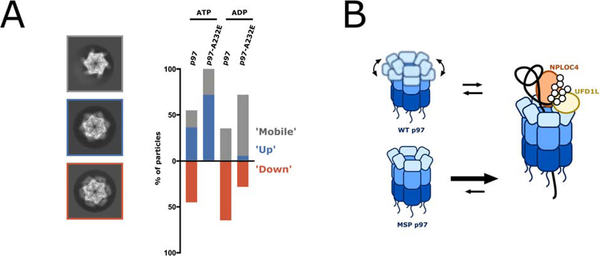
Figure 6. MSP mutations alter NTD conformational dynamics.
(A) Reference-free 2D-class averages from single-particle cryo-EM datasets for p97 and p97-A232E in the presence of ATP or ADP were compared to back projections of the maps for p97 in ADP (EMD-3297) and p97 in ATPγS (EMD-3299), and NTD conformations were assigned to ‘up’ and ‘down’, or ‘mobile’ when NTDs were not resolved. Shown on the left are representative class averages for the ‘mobile’ conformation (top with grey outline, taken from p97-A232E in ATP), the ‘up’ conformation (middle with blue outline, taken from p97 in ATP), and the ‘down’ conformation (bottom with orange outline, taken from p97 in ATP). The proportion of particles in ‘down,’ ‘up,’ and ‘mobile’ conformations are plotted on the right as a percentage of total particles classified. NTDs in the ‘mobile’ conformation either originate from averaging mixed ‘up’ and ‘down’ states in more dynamic parts of the hexamer or from actual mobility after dislodging from the defined ‘down’ state, and are therefore in the bar graph grouped together with the ‘up’ state. See also Figure S4. (B) Model for the increased unfoldase activity of MSP mutant p97. While the NTDs in wild-type hexamers are moving between ‘up’ and ‘down’ conformations dependent on the nucleotide state, the NTDs of MSP-mutant hexamers remain in an ‘up’ conformation, allowing both, faster UN binding and faster substrate processing.