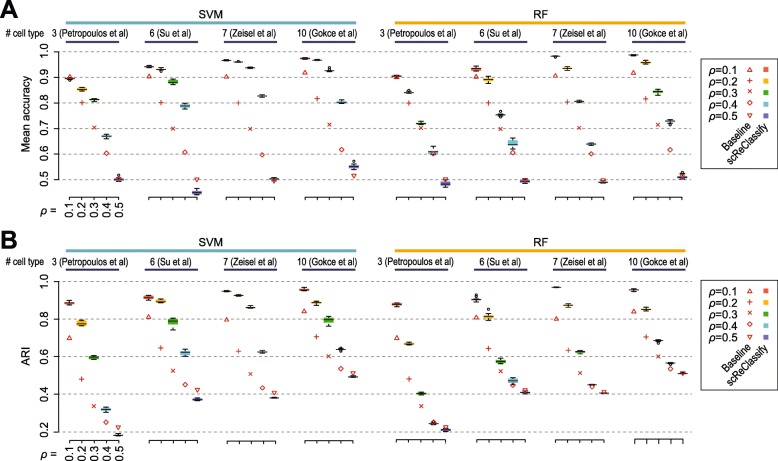
Fig. 4.
Evaluation of scReClassify on experimental datasets. Varying percentages of mislabelled cells were introduced to each scRNA-seq dataset to create an initial cell type annotation with different level of label noise (ρ range from 0.1 to 0.5). The performance in terms of mean accuracy (a) and ARI (b) calculated from the gold standard cell type annotation (annotation of each dataset from its original studies) and the initial cell type annotation (baseline), and the scReClassify corrected cell type annotation. scReClassify was repeated 10 times to capture the variability and shown as boxes coloured according to the percentages of mislabelled cells