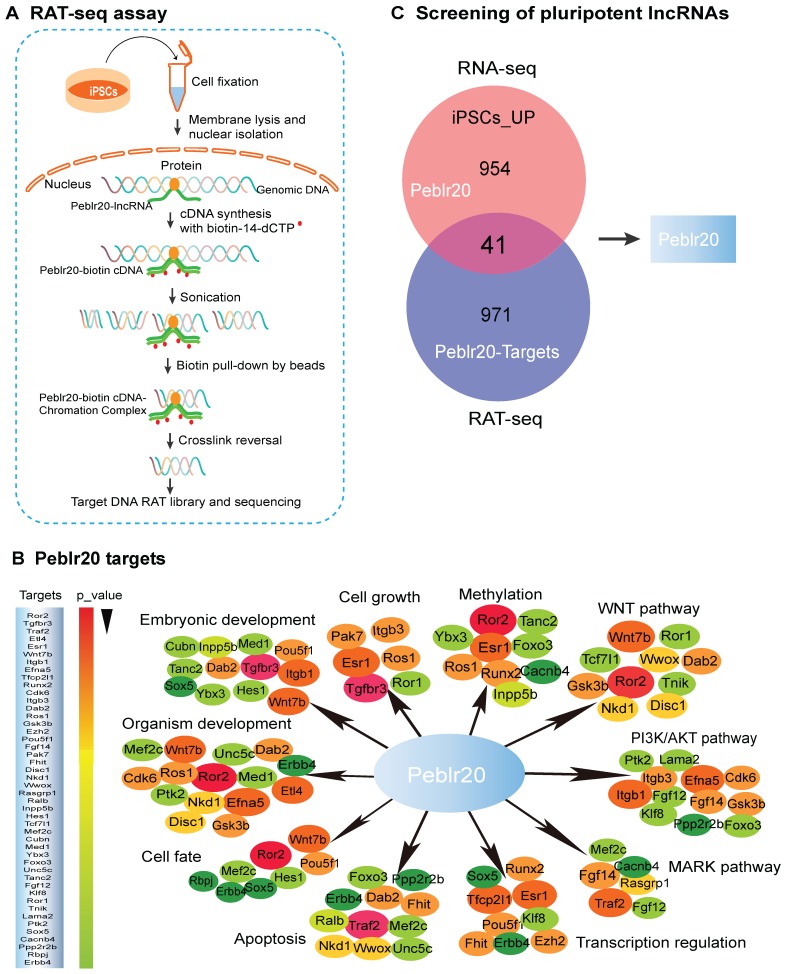
Figure 1.
Identification of pluripotent lncRNAs by RNA-seq and RAT-seq. (A) Schematic diagram of the RNA reverse transcription-associated trap sequencing (RAT-seq) assay. RNA was in situ reverse transcribed using Peblr20 lncRNA-specific antisense oligo primers and biotin-14-dCTP. The biotin- cDNA/chromatin DNA complex was pulled down by biotin-streptavidin magic bead after sonication. The Peblr20-binding chromatin DNAs were isolated for library sequencing. (B) The Peblr20 interaction network targets identified by RAT-seq. Peblr20 binds to many pathway genes that are involved in embryonic development, transcription regulation, cell fate, signaling pathways, etc. Target genes were listed in the order of p-value. (C) Profiling pluripotent lncRNAs by integrating RNA-seq and RAT-seq. The conventional RNA-seq approach identifies thousands of lncRNAs that are differentially expressed during pluripotent reprogramming. A modified RAT-seq approach was performed to map the genome wide interacting target genes for the selected lncRNA. Using this strategy, we identified Peblr20 as a pluripotent lncRNA, because it is activated in iPSCs and binds to multiple stemness gene targets.