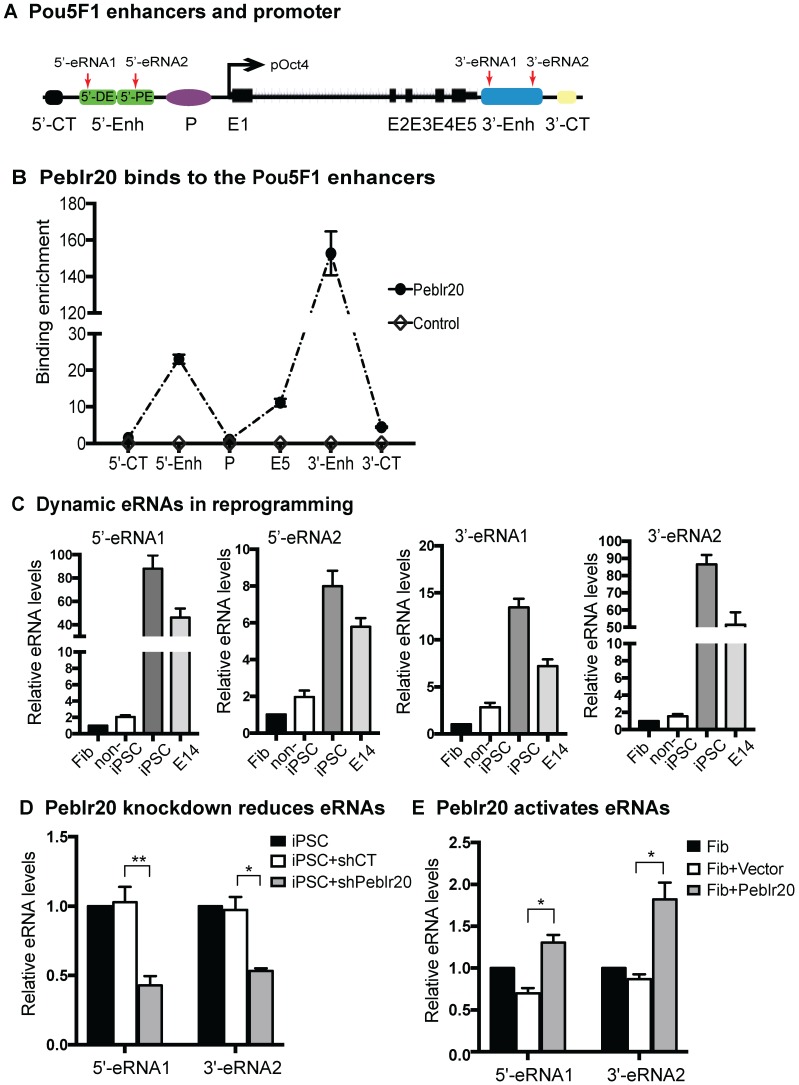
Figure 6.
Peblr20 activates the Pou5F1 enhancer RNA pathway. (A) Location of Pou5F1 enhancer RNAs. 5'-eRNA1: the 5-distal enhancer (5'-DE); 5'-eRNA2: the 5'-proximal enhancer (5'-PE); 3'-eRNA1 and 2: the 3'-enhancer (3'-Enh); P: the Pou5F1 promoter; 5'-CT, 3'-CT: the RAT 5'- and 3'- control sites; E1-E5: Pou5F1 exons 1-5. (B) Peblr20 lncRNA-Pou5F1 DNA interactions by Q-PCR. RAT library samples were used to perform Q-PCR to quantitate binding intensity. The results were normalized to the value of the streptavidin bead pulldown control. Control: the RAT-seq library was constructed using random oligo primers; Obelr20: the RAT-seq library was constructed using Peblr20-speccific primers. (C) Dynamic expression of Pou5F1 eRNAs in reprogramming. Cells were collected at various stages of reprogramming and the expression of eRNAs was measured by RT-Q-PCR. Fib: fibroblast; URC: non-iPSC: fibroblasts that carried the lentiviral OSKM but failed to complete reprogramming; iPSC: induced pluripotent stem cells from secondary MEFs; E14: mouse pluripotent stem cell line as a positive control. Gene expression was normalized to β-Actin internal control. (D) Peblr20 knockdown inhibits Pou5F1 eRNA. iPS+shCT: iPSCs transfected with the random shRNA vector control; iPS +shPeblr20: iPSCs transfected with shPeblr20. Gene expression was normalized to β-Actin control. (E) Peblr20 overexpression activates Pou5F1 enhancer RNAs in fibroblasts. Cells were collected from gain-of-function experiments and eRNAs expressions were measured by RT-Q-PCR. Fib+Vector: fibroblasts transfected with vector control; Fib+Peblr20: fibroblasts overexpressed Peblr20. All experiments were performed in triplicate and statistically significant differences by Student's t test. * P < 0.05; ** < 0.01.