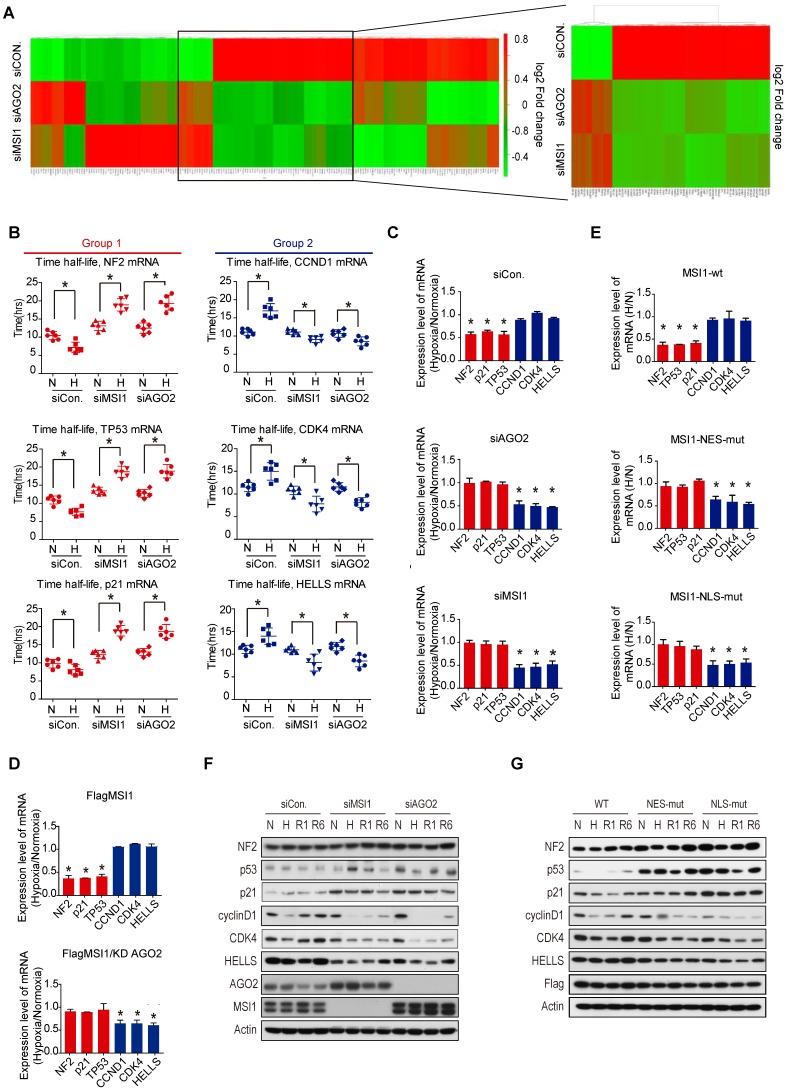
Figure 3.
MSI1/AGO2 mediate the stability of downstream mRNA targets. (A) Parental and MSI1- or AGO2-knockdown cells under normoxia and hypoxia conditions were subjected to a gene expression microarray. Bioinformatics analysis of the microarray data with focus on the 336 common targets of MSI1 and AGO2 identified by RIP-Seq showed the hierarchical clustering these common targets in the heat map. The red and green colors respectively indicate the differentially up or downregulated genes. Each group were done in three distinct biological replicates and the means signals were transformed to the log2 scale. (B) Actinomycin D (Act. D, 5 μg/ml) was added to parental and MSI1- or AGO2-knockdown cells for the indicated times. We compared the half-life distribution of TP53, NF2, CDKN1A, CCND1, CDK4 and HELLS mRNA levels between parental and MSI1- or AGO2-knockdown cells. The RNA expression levels are shown below each the respective box-plots. (C, E) Parential and MSI1- or AGO2-knockdown 05MG cells, as well as MSI1-wt, MSI1-NES-mut, and MSI1-NLS-mut transfected 05MG cells were treated with normoxia or hypoxia conditions for 24 hours. Purified RNA was subjected to RT-PCR with primers specific to TP53, NF2, CDKN1A, CCND1, CDK4 and HELLS. The mRNA levels under hypoxia were normalized by that under normoxia and shown as relative value in the chart. (D) 05MG/MSI1-wt and 05MG/MSI1-wt/shAGO2 cells were treated with normoxia or hypoxia conditions for 24 hours. Purified RNA was subjected to quantitative RT-PCR with primers specific to TP53, NF2, CDKN1A, CCND1, CDK4 and HELLS. The mRNA levels under hypoxia were normalized by that under normoxia and shown as relative value in the chart. * P<0.05. (F) 05MG with AGO2- or MSI1-knockdown, and (G) MSI1-wt, MSI1-NES-mut, or MSI1-NLS-mut overexpression under normoxia or hypoxia for 24 hours, or recovery from hypoxia condition for 1 and 6 hours, were subjected to Western blotting with antibody of NF2, p53, p21, cyclin D1, CDK4, HELLS, AGO2, MSI1, Flag and actin.