1. Introduction
Until recently, alcohol abuse and dependence, as described by the Diagnostic and Statistical Manual of Mental Disorders (DSM IV; (American Psychiatric Association, 2000)), were the most studied problematic outcomes for the clinical consequences of alcohol consumption. As diagnostic measures, alcohol abuse or dependence were restricted to classifying individuals as “affected” or “unaffected” with little sensitivity for underlying profiles of endorsement of the 11 symptoms (7 for dependence and 4 for abuse) that were being used to describe alcohol-related behavior. Hence, especially when used as research outcomes, there was significant concern about DSM-IV diagnoses of abuse or dependence to (1) reflect individual differences with respect to the underlying constructs believed to be represented by the 11 symptoms that research showed is indicative of a single continuum (Hasin and Beseler, 2009, NRC, 2011), and (2) reflect differences in severity across all of the addiction domains (i.e., (a) a compulsion to seek and/or take alcohol, (b) loss of control over alcohol consumption, and (c) emergence of a negative emotional state) captured by the symptoms. Not surprising, recent studies and recommendations which arose out of the Substance Related Disorders Working Group of the DSM-5 taskforce suggested that diagnosis of an alcohol use disorder (AUD) be based on 11 symptoms derived from the integration of the DSM-IV dependence symptoms with three of the DSM-IV abuse criteria (i.e., less ‘recurrent legal problems’) and an alcohol craving criterion. Notably, a continuum should be used to describe AUDs (i.e., unaffected = endorsing 0–1 out of 11 symptoms; mild = endorsing 2–3 out of 11 symptoms; moderate = endorsing 4–5 symptoms out of 11 symptoms, and severe = endorsing 6 or more out of 11 symptoms).
As part of the debate on the utility and suitability of the dichotomous measure of alcohol dependence (AD), as opposed to a continuous measure of alcohol problem severity in genetic research, we recently examined the assumption of genetic homogeneity across all DSM-IV dependence symptoms using genomewide SNP data (Palmer et al., 2015b). Validation of the assumption across the seven symptoms affirmed the utility of a factor score across the indices as much of the observed genetic variance was shared across the comorbid items, suggesting common genetic factors underlie the addiction state (Koob et al., 2014), which is reflected in behavioral symptoms included in DSM-5 AUD (American Psychiatric Association, 2013). The results also indicated that effects observed upon a latent continuum of AD risk (as indicated by DSM-IV dependence symptoms) may not be truly reflective of the entire liability continuum, as there also exists symptom-specific genetic variance that may be imparted by the study of multiple factors (as previously suggested using a multivariate twin study approach (Kendler et al., 2012)). This latter point was recently reflected in a report by Hart and colleagues which showed variation in the association between common genetic variants within the alcohol dehydrogenase gene (ADH1B) and each of the diagnostic symptoms of AD (Hart et al., 2016). In perspective, Hart and colleagues were able to determine that previously observed AD genomewide association study (GWAS) associations (Gelernter et al., 2014), for example, in their subjects of African ancestry (i.e., rs2066702 with AD), were primarily driven by signals specific to phenotypic variation in the symptoms ‘Tolerance’ and ‘Much time spent using/recovering from the effects of alcohol’. This observation is important as phenotype-genotypic associations from GWAS are used to inform gene function studies in tissue/cell culture and/or model organisms.
Altogether, our previous study of DSM-IV symptoms and these recent molecular studies of DSM-IV and DSM-5 AUD underscore the need to characterize the multivariate genetic architecture of DSM-5 AUD symptoms. The present paper uses subjects of European ancestry from the Study of Addiction: Genetics and Environment Consortium to characterize the genetic architecture of the 11 DSM-5 AUD symptoms. It builds upon our previous report by comparing several models that test the assumption of genetic homogeneity across AUD symptoms. The goals of this study were to:
Examine the genomewide additive genetic contribution to DSM-5 AUD symptoms (i.e., former DSM-IV abuse and dependence symptoms along with alcohol craving), and
Determine the most parsimonious model of the additive genetic covariance across DSM-5 AUD symptoms.
2. Materials and Methods
2.1. Sample
Data were drawn from the Study of Addiction: Genetics and Environment (SAGE) (Bierut et al., 2010). Analyses focused on 2596 unrelated individuals (44% male; mean age=38.58 years [standard deviation (SD)=9.80]) of European ancestry, which was confirmed using principal component analysis. All subjects were no more related than second cousins(Palmer et al., 2015a). Additional details on SAGE are available at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1.
2.2. Phenotype
The dependent variables of interest for the current study were DSM-5 AUD symptoms (coded as present or absent) that were approximated from the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (Bucholz et al., 1994, Hesselbrock et al., 1999). Specifically, DSM-IV alcohol abuse and dependence symptoms were extracted from the SSAGA interview and combined with a separate measure of alcohol craving that was also included in the assessment. Craving was defined in the SSAGA as endorsement of the item ‘In situations where you couldn’t drink, did you ever have such a strong desire for it that you couldn’t think of anything else’; respondents who answered yes received a score of 1; ‘No’ was coded as 0; non-users received a missing value (Agrawal et al., 2013). All responses were limited to individuals with a history of exposure to alcohol (and possibly other substances).
2.3. Genotyping and Quality Control
Subjects within SAGE were genotyped using the ILLUMINA Human 1M platform. Quality control of the sample included: (1) removal of non-autosomal SNPs, (2) removal of markers with an allele frequency <1%, (3) exclusion of markers with a call rate less than 98%, and (4) removal of SNPs that show evidence of deviation from Hardy–Weinberg Equilibrium (HWE; p-value < 0.0001) to minimize any possible bias due to assortative mating (Agrawal et al., 2006, Grant et al., 2007). A total of 796,125 autosomal SNPs were carried forward in the analyses. These same SNPs were also used to conduct the aforementioned selection of distantly related EA individuals from the entire set of SAGE participants (N=4121) (i.e., using the software package: Genomewide Complex Trait Analysis [GCTA])(Yang et al., 2010, Lee et al., 2011, Yang et al., 2011).
2.4. Statistical Analysis
The EA sample (N=2596) was used in all parts of the analytical framework, which included (1) development of a phenotypic factor comprised of shared variance among DSM-5 items using randomly selected individuals to create two halves of the sample for exploratory and confirmatory models (conducted in Mplus version 7) (Muthén and Muthén, (1998–2012)), (2) Genomic-relatedness-matrix restricted maximum likelihood (GREML; implemented in GCTA) analysis of the individual symptoms, AUD factor score (standardized [mean=0, standard deviation=1]), AUD diagnosis (i.e., 0=0–1 symptoms, 1=2–11 symptoms), and log-transformed DSM-5 AUD diagnosis severity (i.e., Ln(1+DSM-5 AUD diagnosis [0=0–1 symptoms, 1=mild [2–3 symptoms], 2=moderate[4–5 symptoms], 3=severe [7+ symptoms]]).
2.4.1. Estimation of Additive Genetic Effects
GREML was used to determine the SNP-heritability (h2SNP) of DSM-5 AUD factor score, severity (i.e., mild, moderate, severe), diagnosis (i.e., control vs. case [i.e., 2+ symptoms]), and individual symptoms. This approach was implemented using GCTA. GREML utilizes a genetic relationship matrix to decompose phenotypic variance into genetic effects captured by the common SNPs and error variance. The SNP-heritability estimates were transformed on the liability scale to account for distributional differences in prevalence of AUD and endorsement of AUD symptoms observed in this case/control study versus the general population (i.e. the proportion of cases in this study is higher than what is seen in the population). Lifetime population prevalence estimates that were used to transform the SNP-heritability and co-heritability estimates were calculated for DSM-5 AUD diagnosis and individual symptoms from the National Epidemiologic Survey on Alcohol and Related Conditions (NESARC, wave 1; N=43,093) (Hasin and Grant, 2015) and, for craving from the National Longitudinal Alcohol Epidemiologic Survey (NLAES)(Grant et al., 2003). Craving in NLAES was defined by endorsement of at least one of two possible items: ‘Want to drink so badly that you couldn’t think of anything else’ and ‘Feel a very strong desire or urge to drink’. Prevalence was calculated for individuals in NESARC and NLAES who (1) self-reported non-Hispanic White ethnicity, (2) were aged 18–79 years, and (3) reported lifetime exposure to alcohol (see Table 2). All analyses controlled for gender, age, and the first five ancestral principal components.
Table 2.
Univariate SNP heritability (h2SNP), bivariate correlations, and standard errors (SE)
| Symptom | Population Prevalencea | h2SNP | Inter-symptom genetic correlations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sx1 | Sx2 | Sx3 | Sx4 | Sx5 | Sx6 | Sx7 | Sx8 | Sx9 | Sx10 | Sx11 | ||||
| Sx1 | 4.8 | 0.18 (0.12)b | 1.00 | |||||||||||
| Sx2 | 31.6 | 0.33 (0.20)* | 1.00 (0.88) | 1.00 | ||||||||||
| Sx3 | 11.8 | 0.37 (0.14)** | 1.00 (0.25)* | 0.57 (0.24)b | 1.00 | |||||||||
| Sx4 | 25.5 | 0.34 (0.18)* | 0.87 (0.29)* | 0.81 (0.28)* | 0.53 (0.24)b | 1.00 | ||||||||
| Sx5 | 18.3 | 0.36 (0.23)b | 1.00 (0.44) | 0.71 (0.33)b | 0.98 (0.26)** | 0.46 (0.33) | 1.00 | |||||||
| Sx6 | 23.9 | 0.39 (0.20)* | 1.00 (0.38) | 0.64 (0.27)b | 0.88 (0.21)** | 0.58 (0.27)b | 0.64 (0.32)b | 1.00 | ||||||
| Sx7 | 30.8 | 0.24 (0.18)b | 1.00 (0.47) | 1.00 (0.48) | 0.70 (0.25)b | 1.00 (0.31)* | 0.94 (0.33)* | 1.00 (0.36) | 1.00 | |||||
| Sx8 | 13.6 | 0.13 (0.18) | 1.00 (0.51) | 0.85 (0.56) | 0.75 (0.44)b | 0.49 (0.49)b | 0.65 (0.44) | 0.89 (0.59)b | 0.87 (0.53) | 1.00 | ||||
| Sx9 | 3.7 | 0.28 (0.13)* | 1.00 (0.20)* | 1.00 (0.43) | 0.74 (0.20)* | 0.53 (0.27)b | 0.87 (0.22)* | 0.64 (0.27)* | 0.96 (0.29)* | 0.47 (0.40) | 1.00 | |||
| Sx10 | 11.7 | 0.31 (0.14)* | 1.00 (0.24)* | 0.81 (0.21)* | 0.79 (0.17)** | 0.72 (0.23)* | 0.61 (0.27)b | 0.75 (0.22)* | 0.86 (0.26)* | 0.95 (0.50)* | 0.82 (0.20)* | 1.00 | ||
| Sx11 | 9.9 | 0.24 (0.21) | 0.73 (0.37) | 0.66 (0.42) | 0.54 (0.32) | 0.68 (0.36)b | 1.00 (0.59) | 0.34 (0.42) | 0.38 (0.47) | 0.21 (0.73) | 0.73 (0.28)b | 0.61 (0.34) | 1.00 | |
| Composite phenotypes | ||||||||||||||
| DSM-5 AUD diagnosis | 0.14 (0.21) | |||||||||||||
| DSM-5 AUD severity | 0.22 (0.13)* | |||||||||||||
| DSM-5 AUD factor | 0.36 (0.13)** | |||||||||||||
Notation:
Lifetime population prevalence of symptoms 1 thru 10 were derived from the National Epidemiologic Survey on Alcohol and Related Conditions (NESARC, wave 1; N=43,093); the prevalence for symptom 11 (craving) was derived from National Longitudinal Alcohol Epidemiologic Survey (NLAES). Population prevalence rates were used to transform the SNP-heritability and co-heritability estimates.
- p< 0.10,
- p < 0.05,
- p< 0.01.
Abbreviations:
Sx1: Recurrent use resulting in failure to fulfill major role obligations at work, school, or home
Sx2: Recurrent use in situations in which it is physically hazardous
Sx3: Continued use despite persistent or recurrent social or interpersonal problems caused or exacerbated by the effects of the alcohol
Sx4: Tolerance
Sx5: Withdrawal
Sx6: Taken in larger amounts or over a longer period than was intended
Sx7: Persistent desire or ther are unsuccessful efforts to cut down or control alcohol use
Sx8: A great deal of time is spent in activities necessary to obtain alcohol, use alcohol, or recover from its effects
Sx9: Given up or cut back on important activities in order to drink
Sx10: Continued to use alcohl despite knowledge of having persistent or recurrent physical or psychological problem that is likely to have been caused by the alcohol
Sx11: Craving
2.4.2. Estimation of the Covariance Explained by SNPs
Two multivariate approaches were used to determine whether the same genetic factors contribute to the phenotypic correlation between AUD symptoms: (1) The Common Pathway Model (CPM) and (2) Exploratory Genetic Factor Analysis (EGFA). In addition, the EGFA was followed up with three confirmatory factor analyses to determine the most parsimonious genetic architecture across the criteria. Three multivariate models were tested: (a) a common genetic factor model, (b) a 2- factor model where factor-1 was indicated by the three former DSM-IV abuse symptoms and craving and factor-2 was indicated by the seven former DSM-IV dependence items), and (c) a 2-genetic factor model in which craving was allowed to cross-load across the factors). The CFA models were compared using the Akaike Information Criteria (AIC) (Akaike, 1973).
In the CPM approach, a latent variable representing the shared variance across all symptoms was decomposed into genetic and error variance in two steps. First, an exploratory factor model (EFA) was fitted to a random selected half of the sample to determine the phenotypic factor structure of AUD; this model was then confirmed using confirmatory factor analysis (CFA) of the remaining half of the sample. AUD factor scores were extracted from the full sample and used in the analyses described above. In the EGFA approach, which represents a multivariate extension of GREML, a factor analysis was conducted on the 11×11 variance/covariance matrix of inter-criterion bivariate SNP heritabilities through a series of steps. First, GREML was used to estimate bivariate SNP genetic covariance estimates across each pair of criteria. Next, these estimates were used to construct an 11×11 genetic variance/covariance matrix. Because covariance matrices constructed from bivariate estimates may not be positive definite, we determined the nearest positive definite variance/covariance matrix using the Higham algorithm (Higham, 2002) within the nearPD package in R, version 3.4.0 (Team, 2017). Finally, we conducted factor analysis of the variance/covariance matrix to determine the factor structure of the multivariate genetic relationship between AUD symptoms. To determine the number of genetic factors, we employed Parallel Analysis implemented in R with the nFactors package. This approach has been shown to outperform other methods under a variety of conditions (Ledesma and Valero-Mora, 2007). A factor was retained if the eigenvalue of the genetic variance/covariance matrix was greater than the 95th percentile of the distribution of eigenvalues derived from random data (generated with 1000 iterations). All analyses for the CPM approach were conducted in Mplus and all analyses for the EGFA approach were conducted in R using the OpenMx and Psych packages; bootstrapped confidence intervals for the EGFA path loadings were obtained using 1000 replicates.
3. Results
3.1. Symptom levels and phenotypic covariance in SAGE
The prevalence of endorsement for each AUD symptom and the tetrachoric correlations among all items are presented in Table 1. Approximately 66.49% (n = 1716) of participants met the diagnostic symptoms for DSM-5 AUD. Phenotypic tetrachoric correlations were high, indicating that symptoms frequently co-occur, and ranged from 0.73 – 0.91 (with the lowest correlations being 0.73 between craving and two criteria: tolerance and ‘Taking alcohol in larger amounts or over a longer period than was intended’).
Table 1.
Sample prevalence and associations among DSM-5 alcohol dependence symptoms.
| Sample Prevalence | Tetrachoric Correlation | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symptom | N | % | Sx1 | Sx2 | Sx3 | Sx4 | Sx5 | Sx6 | Sx7 | Sx8 | Sx9 | Sx10 | Sx11 | |
| Sx1 | 904 | 34.8 | 1.00 | |||||||||||
| Sx2 | 1677 | 64.6 | 0.81 | 1.00 | ||||||||||
| Sx3 | 1188 | 45.8 | 0.83 | 0.84 | 1.00 | |||||||||
| Sx4 | 1263 | 48.7 | 0.75 | 0.74 | 0.76 | 1.00 | ||||||||
| Sx5 | 616 | 23.7 | 0.83 | 0.81 | 0.83 | 0.76 | 1.00 | |||||||
| Sx6 | 1597 | 61.5 | 0.77 | 0.79 | 0.80 | 0.75 | 0.74 | 1.00 | ||||||
| Sx7 | 1110 | 42.8 | 0.81 | 0.75 | 0.82 | 0.76 | 0.83 | 0.81 | 1.00 | |||||
| Sx8 | 704 | 27.1 | 0.86 | 0.80 | 0.83 | 0.76 | 0.84 | 0.77 | 0.79 | 1.00 | ||||
| Sx9 | 692 | 26.7 | 0.91 | 0.84 | 0.87 | 0.78 | 0.85 | 0.76 | 0.84 | 0.89 | 1.00 | |||
| Sx10 | 1249 | 48.1 | 0.84 | 0.86 | 0.83 | 0.77 | 0.85 | 0.81 | 0.80 | 0.81 | 0.85 | 1.00 | ||
| Sx11 | 545 | 21.0 | 0.82 | 0.77 | 0.81 | 0.73 | 0.87 | 0.73 | 0.81 | 0.83 | 0.86 | 0.82 | 1.00 | |
Note: Table shows
Sx1: Recurrent use resulting in failure to fulfill major role obligations at work, school, or home
Sx2: Recurrent use in situations in which it is physically hazardous
Sx3: Continued use despite persistent or recurrent social or interpersonal problems caused or exacerbated by the effects of the alcohol
Sx4: Tolerance
Sx5: Withdrawal
Sx6: Taken in larger amounts or over a longer period than was intended
Sx7: Persistent desire or there are unsuccessful efforts to cut down or control alcohol use
Sx8: A great deal of time is spent in activities necessary to obtain alcohol, use alcohol, or recover from its effects
Sx9: Given up or cut back on important activities in order to drink
Sx10: Continued to use alcohol despite knowledge of having persistent or recurrent physical or psychological problem that is likely to have been caused by the alcohol
Sx11: Craving
3.2. Univariate additive genetic effects on DSM-5 AUD diagnosis and symptoms
Common SNPs explained 14% (standard error [SE] = 0.21, p = 0.24 of the variation in DSM-5 AUD diagnosis and 22% (SE = 0.13, p = 0.04) of the variation in AUD severity (i.e., ln-transformed DSM-5 AUD categories). Across the 11 AUD symptoms, SNP-heritability estimates varied from 13% (Great time spent using/recovering) to 39% (Using longer than intended), with five of the 11 items reaching significance (p < 0.05) and four items reaching nominal significance (p < 0.10) (see Table 2).
3.3. Analysis of the genetic covariance across DSM-5 AUD symptoms
CPM approach.
Exploratory and confirmatory analyses of phenotypic data revealed a single latent variable (AUD factor; see Table 3). All items loaded on the single latent factor >0.84. Excellent model fit (RMSEA < 0.05, CFI/TLI > 0.95) supports the CPM that describes the phenotypic relationships between symptoms as arising from an unobserved latent trait. Common SNPs explained 36% (standard error [SE] = 0.13, p = 0.002) of the variation in the AUD factor score.
Table 3.
CPM Approach: Exploratory and confirmatory factor models of alcohol dependence symptoms
| Exploratory | Confirmatory | ||||
|---|---|---|---|---|---|
| Parameters | Sample-1 EFA (n=1298) | Sample-2 CFA (n=1298) | Full Sample CFA (N=2596) | ||
| Fit statistics | |||||
| χ2 | 109.596 | 100.862 | 179.382 | ||
| df | 44 | 44 | 44 | ||
| RMSEA | 0.034 | 0.032 | 0.034 | ||
| CFI | -- | 0.999 | 0.998 | ||
| TLI | -- | 0.998 | 0.998 | ||
| RMSR | 0.026 | -- | -- | ||
| Factor loadings | |||||
| Sx1: Recurrent use resulting in failure to fulfill major roles | 0.927 | 0.921 | 0.924 | ||
| Sx2: Recurrent use in situations in which it is physically hazardous | 0.897 | 0.898 | 0.897 | ||
| Sx3: Continued use despite persistent or recurrent social problems | 0.916 | 0.909 | 0.912 | ||
| Sx4: Tolerance | 0.844 | 0.823 | 0.833 | ||
| Sx5: Withdrawal | 0.907 | 0.927 | 0.917 | ||
| Sx6: Taken in larger amounts or over a longer period than was intended | 0.867 | 0.859 | 0.863 | ||
| Sx7: Persistent desire to cut down or control alcohol use | 0.873 | 0.899 | 0.885 | ||
| Sx8: A great deal of time spent to obtain/use/recover from alcohol | 0.935 | 0.891 | 0.913 | ||
| Sx9: Given up or cut back on important activities in order to drink | 0.952 | 0.956 | 0.954 | ||
| Sx10: Continued to use alcohol despite knowledge physical or psychological problems | 0.926 | 0.909 | 0.917 | ||
| Sx11: Craving | 0.894 | 0.910 | 0.900 | ||
Note: EFA = Exploratory factor analysis; CFA = Confirmatory factor analysis; df = degrees of freedom; RMSEA = Root Mean Square Error Of Approximation; CFI = Comparative Fit Index; TLI = Tucker Lewis Index; RMSR = Root Mean Square Residual
EGFA Approach.
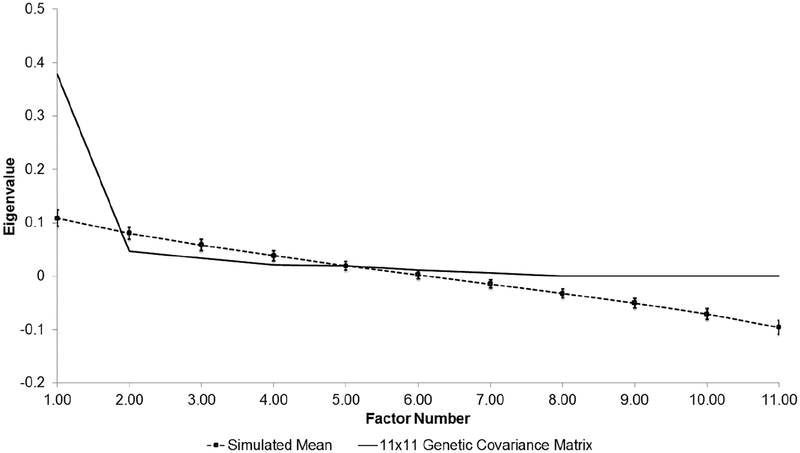
Across AUD symptoms, the pattern of inter-symptom SNP correlations was generally high (strong rG-SNP > 0.60), suggesting shared genetic variance across symptoms (see right side of Table 2). However it is important to recognize that several of these estimates were inflated in instances where the h2SNP of at least one of the symptoms was non-significant (i.e., only a small proportion of the phenotypic variance in the DSM-5 criterion is explained by genetic variation). Analysis of the 11×11 genetic variance/covariance matrix suggested a single genetic factor parsimoniously describes much of the shared genetic variance across the 11 criteria (see Supplementary Table S1 for the genetic variance/covariance matrix). Parallel analysis indicated that the first eigenvalue derived from this matrix exceeded the 95th percentile of the distribution of eigenvalues derived from random data (see Figure 1). Genetic factor loadings for AUD symptoms were high (>0.60) and the total genetic variance of each criteria attributable to the factor ranged from 38% for ‘craving’ to 89% for ‘failure to fulfill major roles’ (see Table 4 for a summary of factor loadings and percent variance explained in the EGFA). Our analysis of competing models of additive genetic effects on AUD indicated that the model containing a single genetic factor provided the best fit to the data (χ2=49905.53, degrees of freedom (df)=44, AIC=49817.53). On the contrary, the two-factor genetic model that allowed for a correlation between the genetic factors was less parsimonious (compared to the one-factor model: ΔAIC=4046.55), but estimated the correlation between the abuse and dependence factors at 1.00 [95% confidence interval=0.99, 1.00]. We also examined a bi-genetic factor model that allowed craving to cross-load across the factors; results were similar and this adjusted model fit the data slightly worse (compared to the one-factor model: ΔAIC=4048.55). Overall, both the EGFA and CFA suggest shared additive genetic effects across symptoms of DSM-5 AUD.
Figure 1. Parallel analysis of 11×11 genetic covariance matrix for DSM-5 AUD symptoms.
Observed eigenvalues (solid line) are compared to 95 percentile of the eigenvalue distribution (dashed line [with standard error]) derived from 1000 randomly generated datasets. All factors left of where the solid lines first intersects with the dashed line are retained and their effects described in Table 4.
Table 4.
EGFA Approach: Genetic variance in each DSM-5 symptom explained by common genetic factor.
| Symptom | Factor Loading [95% CI] | % Total Genetic Variance Explained | |
|---|---|---|---|
| Sx1: Recurrent use resulting in failure to fulfill major roles | 0.93 [0.92,0.94] | 87% | |
| Sx2: Recurrent use in situations in which it is physically hazardous | 0.81 [0.80,0.83] | 66% | |
| Sx3: Continued use despite persistent or recurrent social problems | 0.86 [0.85,0.87] | 74% | |
| Sx4: Tolerance | 0.74 [0.73,0.76] | 55% | |
| Sx5: Withdrawal | 0.80 [0.79,0.82] | 64% | |
| Sx6: Taken in larger amounts or over a longer period than was intended | 0.83 [0.82,0.84] | 69% | |
| Sx7: Persistent desire to cut down or control alcohol use | 0.90 [0.89,0.91] | 81% | |
| Sx8: A great deal of time spent to obtain/use/recover from alcohol | 0.76 [0.75,0.78] | 58% | |
| Sx9: Given up or cut back on important activities in order to drink | 0.85 [0.84,0.86] | 72% | |
| Sx10: Continued to use alcohol despite knowledge physical or psychological problems | 0.91 [0.90, 0.92] | 83% | |
| Sx11: Craving | 0.61 [0.59, 0.64] | 37% | |
Table showing standardized factor loadings of the exploratory genetic factor analysis along with 95% confidence intervals and squared standardized factor loading (i.e., percent of genetic variance explained).
4. Discussion
This study examined the expanded definition of diagnostic criteria contributing to AUD as defined by the American Psychiatric Association’s DSM-5. Additive genetic effects are partially shared across DSM-5 symptoms of AUD, with genetic correlations > 0.80 for several criteria. However, correlations across some criteria were as low as 0.21 (e.g., craving and time spent), suggesting the possibility of a violation of the assumption of genetic homogeneity underlying the AUD phenotype, but this was not a common occurrence (i.e. percentage of correlations >0.3=98%, >0.6=78%, and >0.8=42%). Notably, while the standard errors for some of these correlation estimates was fairly large (e.g., rG=0.21 (SE=0.73) for the association between craving and ‘A great deal of time spent to obtain/use/recover from alcohol’), other correlations were more precise (e.g., rG=1.00 (SE=0.20) for ‘Recurrent use resulting in failure to fulfill major roles’ associated with ‘Given up or cut back on important activities in order to drink’), and thus, the hypothesis of underlying genetic homogeneity cannot be fully rejected. However, it is possible that the ability to localize genetic loci for AUDs is likely to be reduced when using scoring methods that ignore the fact that symptoms are influenced by shared and non-shared genetic factors (i.e., just as there are a multitude of symptom profiles that lead to an AUD diagnosis, the respective genetic risk profiles for these various symptom profiles may vary accordingly).
The incorporation of previous DSM-IV abuse symptoms and craving for alcohol enhanced the definition of problematic alcohol use. Estimated effects of genetic variation on the newly added alcohol symptoms ranged from 0.10–0.37. Notably, the SNP-heritability of DSM-5 AUD factor was similar to what was previously reported for DSM-IV alcohol dependence as a factor score (Palmer et al., 2015b, Brick et al., 2017). Moreover, our examinations of the genetic influences on the 11 symptoms supports the underlying assumptions of (1) a single underlying dimension of risk that is captured by the symptoms, and (2) common genetic pathways that contribute to ‘craving’, ‘using longer than intended’, ‘withdrawal’, and the other symptoms. These findings align with previous examinations of alcohol symptoms in genetically informed samples, which suggested a single underlying latent trait that is polygenic in nature. It was for this reason that we opted to use Genomic Restricted maximum likelihood (GREML) to understand the relationship among the 11 AUD symptoms because our sample sizes precluded the use of genomewide association analysis which would have resulted in biased SNP-estimates that reflect only a small portion of the phenotypic heritability (i.e., missing heritability; (Manolio et al., 2009)). In the current analysis, we were interested in quantifying the heritability and co-heritability due to common variation. In GREML, there is less emphasis on detecting the small effects of the common variants, but instead more emphasis on aggregated effects. Our examination of alternative factorial configurations of the criteria (i.e., correlated abuse and dependence genetic factors) provides novel evidence supporting the assumption that the genetic architecture across AUD symptoms is largely shared. Indeed, as the data suggest, GWAS aimed at the factors of tolerance, loss of control and withdrawal (Kendler et al., 2011) may yield loci distinct from those identified using a unidimensional factor score or diagnosis, because of limited power to detect such specificity and also because of AUD-symptom-specific genetic variance. As such, future works should consider analyzing AUD measures in their various forms.
Using these approaches, for the first time we report on the genomewide SNP-heritability of alcohol craving. Similar to our earlier report using a larger, but ancestrally mixed SAGE sample (Agrawal et al., 2013), 21% percent of the total sample endorsed alcohol craving. The univariate SNP-heritability of 0.24 (SE=0.21; post-hoc power=0.57) for craving did not meet our criteria for statistical significance, however, the high loading (>0.85) of the craving item on the latent AUD factor, which had a SNP-heritability of 0.36 (standard error [SE] = 0.13), suggests genetic effects on craving. Notably, our analysis of the genetic effects across symptoms suggested some differential effects of genomewide SNPs across AUD symptoms, but craving was least explained by the common genetic factors. A review of the alcohol literature identified two studies supporting the role of variation across the alpha-synuclein gene (SNCA) and alcohol craving. α-synuclein has been shown to play a role in dopamine functioning across several regions of the brain (e.g., inhibiting dopamine synthesis; (Perez et al., 2002)) – making it a candidate for addiction research. In regards to alcohol, Foroud et al. (Foroud et al., 2007), identified haplotypes of SNPs in SNCA that were associated with alcohol craving, but not DSM-IV alcohol dependence diagnosis, supporting our argument here that the study of individual symptoms for AUD may at times point to sources of liability that may be overlooked when studying only the shared variance across AUD symptoms. More recently, our analysis of genes in the dopamine pathway (i.e., DRD1, DRD2, DRD3, DRD4, SLC6A3, as well as SNCA) also suggested common and specific effects from variants in these genes across craving and alcohol dependence (i.e., without craving) (Agrawal et al., 2013). It is important to note that for the current analyses, craving was assessed using a single item (strong desire to use so couldn’t think of anything else). Prior work has contrasted the contributions of varying definitions of craving on AUD diagnosis (Keyes et al., 2011); the NESARC includes two items that effectively separate “strong desire or urge” from “couldn’t think about anything else” with the former being more commonly reported in population samples than the latter. However, “strong desire” is incorporated into the International Classification of Disease (ICD-10) diagnosis of AUD (Yoshimura et al., 2016) and is part of the phrasing of the corresponding DSM-5 item. Therefore, our single item adequately captures the phenomenon of craving, although future studies might wish to explore alternative conceptualizations of the construct.
The SNP-heritability of DSM-5 AUD diagnosis (h2SNP=0.14) was not significant but approximates the estimate of 0.09 from a recent GWAS meta-analysis of DSM-IV alcohol dependence (Sanchez-Roige et al., 2018). In contrast, SNP-heritability for the AUD factor score (0.36) and for the AUD severity score (0.22) were significant and higher. The use of a quantitative index of liability likely increased sensitivity to capture genetic effects and supports the transition from a binary (DSM-IV) to categorical (DSM-5) definition of AUD. However, the modest reduction in heritability when using the AUD severity score, which represents DSM-5 categories implies some compression of meaningful variability that the full spectrum of scores affords. Indeed, a symptom count was used amongst the few successful GWAS of alcohol dependence (Gelernter et al., 2014). As a symptom count is similar to a unidimensional factor score, our analyses support the use of such symptom counts, potentially augmented to even include indices of alcohol consumption (Saha et al., 2007), as well as other comorbid internalizing and externalizing type disorders that have evidenced genetic overlap (Cerda et al., 2010).
Important considerations for the current study were our inability to model dominance and epistatic effects from genomewide loci, which is a growing area of interest in the field of psychiatric genetics, but is an approach that was not feasible with our current sample size. As such, readers should interpret these effects as the cumulative/additive effect of genomewide SNPs, which is akin to the additive genetic variance component (A) typically examined in the twin literature and that largely contributes to the correlation among relatives. Likewise, an examination of gender was prohibited due to sample size, but twin studies that have explored gender differences (i.e., qualitative and quantitative) have been mixed (Verhulst et al., 2015). Another important consideration to arise from this study is, the varied h2SNP effect sizes across symptoms suggest that larger samples are needed to study individual symptoms with limited (<0.30) genetic effects, particularly if the variance across the set of SNPs influencing each symptom is not constant. We have cautiously interpreted our study findings because of the limited power to detect modest SNP-heritability estimates and genetic correlations, especially in instances where the SNP-heritability of a pair of items was low and non-significant. As is the case with twin and family studies (Verhulst, 2017), cautious interpretation is warranted when estimating and interpreting genetic correlations between phenotypes when the magnitude of the genetic effect is limited. To aid in our interpretation of these data we modeled the raw genetic variance/covariance matrix to minimize bias. Similarly, we compared several multivariate factor models of the genetic covariance matrix and report bootstrapped confidence intervals of the loadings from the most parsimonious model. Altogether, when considering these factor, the pattern of results provide preliminary evidence to suggest that studying the shared liability across all of the DSM symptoms is a more genetically sensitive (i.e., evidencing a moderate heritability [0.30–0.60]) and parsimonious phenotype, since the loci likely reflects the lowest common denominator/factors for AUD.
In conclusion, we discovered that the APA’s DSM-5 definition of alcohol-related problems is a heritable phenotype with varying genetic effects across the individual symptoms with both shared and non-shared genetic variance between them. Though tentative and in need of replication in larger samples, these findings lend support to the use of composite scores, such as factor scores or symptom count as phenotype, as well as the application of genomic structural equation model methods in future studies.
Supplementary Material
Acknowledgements
COGA: The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks); Department of Biomedical and Health Informatics, The Children’s Hospital of Philadelphia; Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA (L. Almasy), Virginia Commonwealth University (D. Dick), Icahn School of Medicine at Mount Sinai (A. Goate), and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); J. McClintick, L. Wetherill, X. Xuei, Y. Liu, D. Lai, S. O’Connor, M. Plawecki, S. Lourens (Indiana University); G. Chan (University of Iowa; University of Connecticut); J. Meyers, D. Chorlian, C. Kamarajan, A. Pandey, J. Zhang (SUNY Downstate); J.-C. Wang, M. Kapoor, S. Bertelsen (Icahn School of Medicine at Mount Sinai); A. Anokhin, V. McCutcheon, S. Saccone (Washington University); J. Salvatore, F. Aliev, B. Cho (Virginia Commonwealth University); and Mark Kos (University of Texas Rio Grande Valley). A. Parsian and H. Chen are the NIAAA Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Role of funding sources
This body of work was supported by research grants from the National Institute on Alcohol Abuse and Alcoholism (K01AA021113 awarded to Rohan Palmer, R01AA025593 awarded to John McGeary and P60AA11998 awarded to Andrew Heath, and K02DA032573 and R21AA021235 awarded to Arpana Agrawal), the National Institute on Drug Abuse (DP1DA04213 awarded to Palmer), and the National Institute on Mental Health (R01MH100141 awarded to Matthew Keller and R56MH108650 awarded to John McGeary). Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The views expressed in this article do not necessarily reflect the position or policy of the Department of Veterans Affairs.
Footnotes
Conflict of interest
All of the authors declare they have co conflicts.
REFERENCES
- AGRAWAL A, HEATH AC, GRANT JD, PERGADIA ML, STATHAM DJ, BUCHOLZ KK, MARTIN NG & MADDEN PA 2006. Assortative mating for cigarette smoking and for alcohol consumption in female Australian twins and their spouses. Behav Genet, 36, 553–66. [DOI] [PubMed] [Google Scholar]
- AGRAWAL A, WETHERILL L, BUCHOLZ KK, KRAMER J, KUPERMAN S, LYNSKEY MT, NURNBERGER JI JR., SCHUCKIT M, TISCHFIELD JA, EDENBERG HJ, FOROUD T & BIERUT LJ 2013. Genetic influences on craving for alcohol. Addict Behav, 38, 1501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AKAIKE H 1973. Information theory and an extension of the maximum likelihood principle”, in Petrov BN; Csáki F, 2nd International Symposium on Information Theory, Tsahkadsor, Armenia, USSR, September 2–8, 1971, Budapest: Akadémiai Kiadó, pp. 267–281. [Google Scholar]
- AMERICAN PSYCHIATRIC ASSOCIATION 2000. Diagnostic and statistical manual of mental disorders (4th ed., text rev.). [Google Scholar]
- AMERICAN PSYCHIATRIC ASSOCIATION 2013. Diagnostic and statistical manual of mental disorders (5th ed.). Washington, DC: Author. [Google Scholar]
- BIERUT LJ, AGRAWAL A, BUCHOLZ KK, DOHENY KF, LAURIE C, PUGH E, FISHER S, FOX L, HOWELLS W, BERTELSEN S, HINRICHS AL, ALMASY L, BRESLAU N, CULVERHOUSE RC, DICK DM, EDENBERG HJ, FOROUD T, GRUCZA RA, HATSUKAMI D, HESSELBROCK V, JOHNSON EO, KRAMER J, KRUEGER RF, KUPERMAN S, LYNSKEY M, MANN K, NEUMAN RJ, NOTHEN MM, NURNBERGER JI JR., PORJESZ B, RIDINGER M, SACCONE NL, SACCONE SF, SCHUCKIT MA, TISCHFIELD JA, WANG JC, RIETSCHEL M, GOATE AM & RICE JP 2010. A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences of the United States of America, 107, 5082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRICK LA, KELLER MC, KNOPIK VS, MCGEARY JE & PALMER RHC 2017. Shared additive genetic variation for alcohol dependence among subjects of African and European ancestry. Addict Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCHOLZ KK, CADORET R, CLONINGER CR, DINWIDDIE SH, HESSELBROCK VM, NURNBERGER JI JR., REICH T, SCHMIDT I & SCHUCKIT MA 1994. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of studies on alcohol, 55, 149–58. [DOI] [PubMed] [Google Scholar]
- CERDA M, SAGDEO A, JOHNSON J & GALEA S 2010. Genetic and environmental influences on psychiatric comorbidity: a systematic review. J Affect Disord, 126, 14–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOROUD T, WETHERILL LF, LIANG T, DICK DM, HESSELBROCK V, KRAMER J, NURNBERGER J, SCHUCKIT M, CARR L, PORJESZ B, XUEI X & EDENBERG HJ 2007. Association of alcohol craving with alpha-synuclein (SNCA). Alcoholism, clinical and experimental research, 31, 537–45. [DOI] [PubMed] [Google Scholar]
- GELERNTER J, KRANZLER HR, SHERVA R, ALMASY L, KOESTERER R, SMITH AH, ANTON R, PREUSS UW, RIDINGER M, RUJESCU D, WODARZ N, ZILL P, ZHAO H & FARRER LA 2014. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Molecular psychiatry, 19, 41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRANT BF, DAWSON DA, STINSON FS, CHOU PS, KAY W & PICKERING R 2003. The Alcohol Use Disorder and Associated Disabilities Interview Schedule-IV (AUDADIS-IV): reliability of alcohol consumption, tobacco use, family history of depression and psychiatric diagnostic modules in a general population sample. Drug Alcohol Depend, 71, 7–16. [DOI] [PubMed] [Google Scholar]
- GRANT JD, HEATH AC, BUCHOLZ KK, MADDEN PA, AGRAWAL A, STATHAM DJ & MARTIN NG 2007. Spousal concordance for alcohol dependence: evidence for assortative mating or spousal interaction effects? Alcohol Clin Exp Res, 31, 717–28. [DOI] [PubMed] [Google Scholar]
- HART AB, LYNCH KG, FARRER L, GELERNTER J & KRANZLER HR 2016. Which alcohol use disorder criteria contribute to the association of ADH1B with alcohol dependence? Addict Biol, 21, 924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HASIN DS & BESELER CL 2009. Dimensionality of lifetime alcohol abuse, dependence and binge drinking. Drug Alcohol Depend, 101, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HASIN DS & GRANT BF 2015. The National Epidemiologic Survey on Alcohol and Related Conditions (NESARC) Waves 1 and 2: review and summary of findings. Soc Psychiatry Psychiatr Epidemiol, 50, 1609–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HESSELBROCK M, EASTON C, BUCHOLZ KK, SCHUCKIT M & HESSELBROCK V 1999. A validity study of the SSAGA--a comparison with the SCAN. Addiction, 94, 1361–70. [DOI] [PubMed] [Google Scholar]
- HIGHAM N 2002. Computing the nearest correlation matrix - a problem from finance. IMA Journal of Numerical Analysis, 22, 329–343. [Google Scholar]
- KENDLER KS, AGGEN SH, PRESCOTT CA, CRABBE J & NEALE MC 2012. Evidence for multiple genetic factors underlying the DSM-IV criteria for alcohol dependence. Mol Psychiatry, 17, 1306–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENDLER KS, KALSI G, HOLMANS PA, SANDERS AR, AGGEN SH, DICK DM, ALIEV F, SHI J, LEVINSON DF & GEJMAN PV 2011. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin Exp Res, 35, 963–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEYES KM, KRUEGER RF, GRANT BF & HASIN DS 2011. Alcohol craving and the dimensionality of alcohol disorders. Psychol Med, 41, 629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOOB GF, BUCK CL, COHEN A, EDWARDS S, PARK PE, SCHLOSBURG JE, SCHMEICHEL B, VENDRUSCOLO LF, WADE CL, WHITFIELD TW JR. & GEORGE O 2014. Addiction as a stress surfeit disorder. Neuropharmacology, 76 Pt B, 370–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEDESMA RD & VALERO-MORA P 2007. Determining the Number of Factors to Retain in EFA: an easy-to-use computer program for carrying out Parallel Analysis Practical Assessment, Research & Evaluation, 12. [Google Scholar]
- LEE SH, WRAY NR, GODDARD ME & VISSCHER PM 2011. Estimating missing heritability for disease from genome-wide association studies. American journal of human genetics, 88, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANOLIO TA, COLLINS FS, COX NJ, GOLDSTEIN DB, HINDORFF LA, HUNTER DJ, MCCARTHY MI, RAMOS EM, CARDON LR, CHAKRAVARTI A, CHO JH, GUTTMACHER AE, KONG A, KRUGLYAK L, MARDIS E, ROTIMI CN, SLATKIN M, VALLE D, WHITTEMORE AS, BOEHNKE M, CLARK AG, EICHLER EE, GIBSON G, HAINES JL, MACKAY TF, MCCARROLL SA & VISSCHER PM 2009. Finding the missing heritability of complex diseases. Nature, 461, 747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUTHÉN LK & MUTHÉN BO (1998–2012). MPlus User’s Guide, Los Angeles, CA, Muthén and Muthén. [Google Scholar]
- NRC 2011. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington (DC). [PubMed] [Google Scholar]
- PALMER RH, BRICK L, NUGENT NR, BIDWELL LC, MCGEARY JE, KNOPIK VS & KELLER MC 2015a. Examining the role of common genetic variants on alcohol, tobacco, cannabis and illicit drug dependence: genetics of vulnerability to drug dependence. Addiction, 110, 530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALMER RH, MCGEARY JE, HEATH AC, KELLER MC, BRICK LA & KNOPIK VS 2015b. Shared additive genetic influences on DSM-IV criteria for alcohol dependence in subjects of European ancestry. Addiction, 110, 1922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREZ RG, WAYMIRE JC, LIN E, LIU JJ, GUO F & ZIGMOND MJ 2002. A role for alpha-synuclein in the regulation of dopamine biosynthesis. J Neurosci, 22, 3090–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAHA TD, STINSON FS & GRANT BF 2007. The role of alcohol consumption in future classifications of alcohol use disorders. Drug Alcohol Depend, 89, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANCHEZ-ROIGE S, PALMER AA, FONTANILLAS P, ELSON S, ADAMS MJ, HOWARD DM, EDENBERG HJ, DAVIES G, CRIST RC, DEARY I, MCINTOSH AM & CLARKE TK 2018. Genome-wide association study meta-analysis of the Alcohol Use Disorder Identification Test (AUDIT) in two population-based cohorts (N=141,958). bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEAM RC 2017. R: A language and environment for statistical computing R Foundation for Statistical Computing Vienna, Austria. [Google Scholar]
- VERHULST B 2017. A Power Calculator for the Classical Twin Design. Behav Genet, 47, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VERHULST B, NEALE MC & KENDLER KS 2015. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med, 45, 1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG J, BENYAMIN B, MCEVOY BP, GORDON S, HENDERS AK, NYHOLT DR, MADDEN PA, HEATH AC, MARTIN NG, MONTGOMERY GW, GODDARD ME & VISSCHER PM 2010. Common SNPs explain a large proportion of the heritability for human height. Nat Genet, 42, 565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG J, LEE SH, GODDARD ME & VISSCHER PM 2011. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet, 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIMURA A, KOMOTO Y & HIGUCHI S 2016. Exploration of Core Symptoms for the Diagnosis of Alcohol Dependence in the ICD-10. Alcohol Clin Exp Res, 40, 2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.