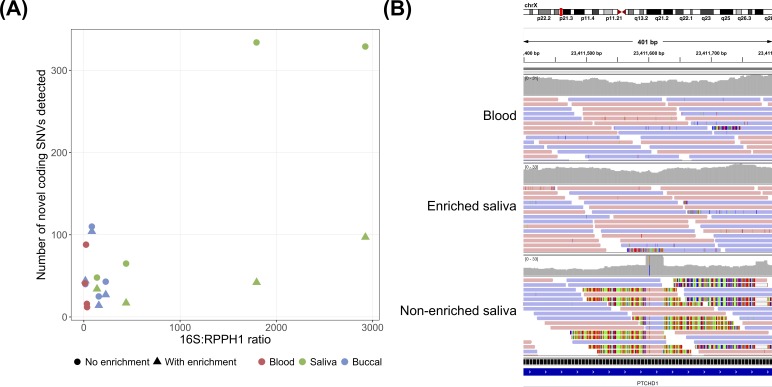
Figure 3.
Bacterial contamination and the detection of false single nucleotide variants (SNVs). (A) Relationship between bacterial DNA concentration and the number of novel coding SNVs detected in each sample. For further details, see figure 2. (B) Integrative Genomics Viewer read pile-up showing a false SNV in an exon of PTCHD1 detected in the non-enriched saliva sample from individual PGPC-0050, but not in the enriched saliva sample or blood sample from the same individual. The false SNV was detected because many short segments of bacterial reads containing a sequence difference relative to the human reference genome aligned to this region. A BLAST search suggested that the aligned bacterial reads were derived from the genome of Fusobacterium periodonticum (99% query cover, 97% identity), a bacterium known to be found in the human oral cavity.45