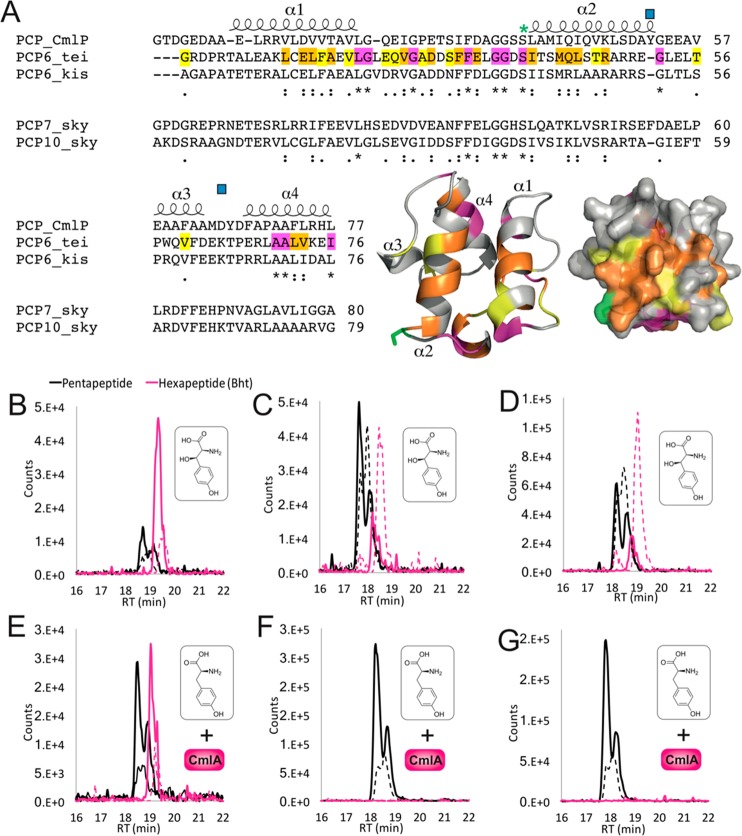
Figure 6.
Investigating CmlA specificity for alternate PCP domains. (A) Sequence alignments of PCP domains from chloramphenicol biosynthesis with PCP domains accepted as CmlA substrates (from the teicoplanin and kistamicin NRPS module 6) and those not accepted by CmlA (from the skyllamycin NRPS modules 7 and 10). Predicated PCP secondary structure is shown, with the degree of homology highlighted in yellow (similar), orange (highly similar), and pink (identical). The post-translationally modified serine residue (green asterisk) and residues implicated in the PCP-acceptance in skyllamycin biosynthesis (blue squares) are indicated. A structural model of PCP6tei demonstrates the location of similar residues on the structure of such a PCP domain (colors as previously indicated). (B–G) LCMS analysis of the reconstitution of hexapeptide extension using separated M5 modules from teicoplanin biosynthesis with hybrid module 6 constructs combining the A-domain from teicoplanin biosynthesis together with PCP domains from the NRPS machinery from kistamicin biosynthesis (module 6, B/E) and skyllamycin biosynthesis (module 7, C/F; module 10, D/G). Peptide biosynthesis was reconstituted with either Bht (upper panels) or Tyr + CmlA (lower panels). Solid lines indicate methylamide peptides (PCP-bound), and dashed lines indicate hydrolyzed peptides (pentapeptide, black line; Bht-containing hexapeptide, pink line).