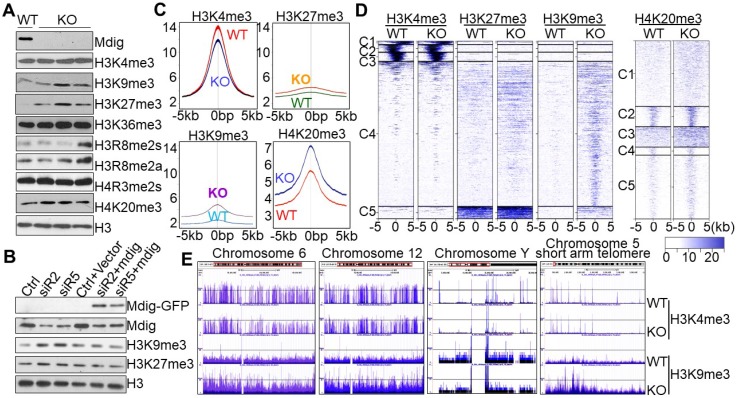
Figure 1.
Knockout of mdig up-regulates repressive histone methylation markers. A. Examination of the methylation states of the indicated lysine markers in histone H3 of the BEAS-2B cells. WT: the cells subjected to CRISPR-cas9-based mdig targeting but failed in mdig knockout; KO: the cells with successful mdig knockout through CRISPR-cas9 approach. Data are representative of one of the two WT colonies and three of six KO colonies. B. Mdig silencing and rescue experiment. The BEAS-2B cells were transfected with the indicated mdig siRNAs alone or in combination with the GFP-tagged mdig expressing vector. C. Histone methylation profiles of ChIP-seq for H3K4me3, H3K27me3, H3K9me3, and H4K20me3 between WT and mdig KO cells. Average plots of the merged peak regions were shown. D. Heatmaps of the merged peak distribution of H3K4me3, H3K27me3, H3K9me3, and H4K20me3 between WT and mdig KO cells. E. All chromosomes showed an enrichment of H3K9me3 following mdig knockout (KO). Representative enrichment diagrams of H3K9me3 are shown for chromosomes 6, 12, Y, and the short arm telomere region of chromosome 5 between WT and KO cells