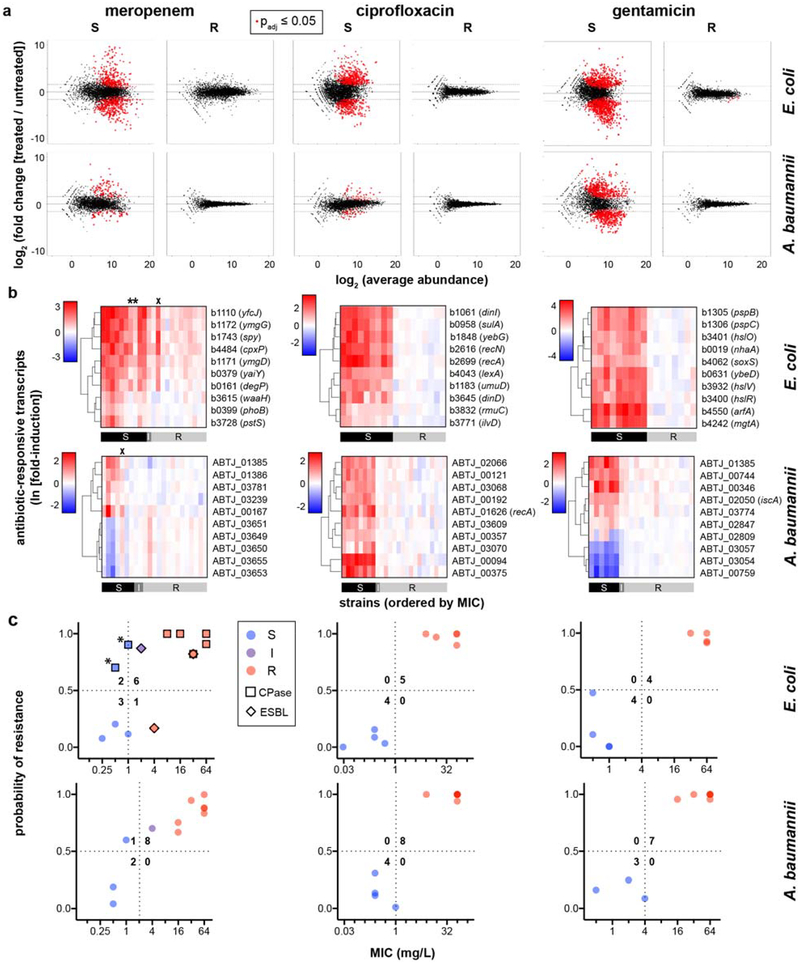
Extended Data Figure 1. Differential gene expression upon antibiotic exposure distinguishes susceptible and resistant strains.
(a) RNA-Seq data from two susceptible (left panels) or two resistant (right panels) clinical isolates of E. coli or A. baumannii treated with meropenem (60 min), ciprofloxacin (30 min), or gentamicin (60 min) at CLSI breakpoint concentrations are presented as MA plots. Statistical significance was determined by a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing, using the DESeq2 package47. (b) Heatmaps of normalized, log-transformed fold-induction of top 10 antibiotic-responsive transcripts from 24 clinical isolates of E. coli or A. baumannii treated at CLSI breakpoint concentrations with meropenem, ciprofloxacin, or gentamicin. Gene identifiers are listed at right, along with gene names if available. CLSI classifications of each strain based on broth microdilution are shown below. * = strains with large inoculum effects in meropenem MIC; x = strains discordant by more than one dilution. (c) GoPhAST-R predictions of probability of resistance from a random forest model trained on NanoString data from the derivation cohort and tested on the validation cohort (y-axis) are compared with standard CLSI classification based on broth microdilution MIC (x-axis) for E. coli (top) or A. baumannii isolates treated with meropenem, ciprofloxacin, and gentamicin. Horizontal dashed lines indicate 50% probability of resistance. Vertical dashed lines indicate the CLSI breakpoint between susceptible and not susceptible (i.e. intermediate/resistant). Numbers in each quadrant indicate concordant and discordant classifications between GoPhAST-R and broth microdilution. Carbapenemase (square outline) and select ESBL (diamond outline) gene content as detected by GoPhAST-R are also displayed on the meropenem plot.